Lecture 6
Data Transformation with R
September 29, 2025
Reading a Data Table as a data.frame
CSV Files
- A CSV (comma-separated values) file is a plain text file where each value is separated by a comma.
- CSV files are widely used for storing data from spreadsheets and databases.
- Example
Absolute Pathnames
- An absolute pathname tells the computer the exact location of a file, starting from the very top folder of your computer.
- This location never changes, no matter where you are working in R.
- This location never changes, no matter where you are working in R.
- In R, you can see the working directory — the folder where R is currently “looking” for files — by running
getwd()in the Console.- In a Posit Cloud, the working directory is
/cloud/project/
- In a Posit Cloud, the working directory is
- Examples of an absolute pathname for
custdata_rev.csv:- On a Mac:
/Users/user/documents/data/custdata_rev.csv
- On Windows:
C:\\Users\\user\\Documents\\data\\custdata_rev.csv- Note: In Windows, we use double backslashes (
\\) because a single backslash (\) is treated as a special character in R.
- Note: In Windows, we use double backslashes (
- On a Mac:
Relative Pathnames
- A relative pathname specifies the location of a file relative to the working directory.
- Examples of a relative pathname for
custdata_rev.csv:- Absolute pathname:
/cloud/project/data/custdata_rev.csv
- Working directory:
/cloud/project/
- Relative pathname:
data/custdata_rev.csv
- Absolute pathname:
- In Posit Cloud, we typically use relative pathnames to access files inside the project folder.
Steps for Reading a CSV File as a data.frame
- Download
custdata_rev.csvfrom the Class Files module in Brightspace.- The file will usually be saved in your computer’s Downloads folder.
- Do not open this file in a spreadsheet app (e.g., Excel or Numbers).
- In Posit Cloud, create a subfolder named data (Files Pane → + Folder).
- Click the data folder, then upload the
custdata_rev.csvfile into it.- At the top of the Files Pane, click Upload to choose and add your file.
- Read the file using a relative pathname:
view(DF)(orView(DF)) opens aDFin a spreadsheet-like viewer.
Load a CSV File Directly from the Web into R
# Read the CSV file directly from the web (GitHub repo)
custdata_web <- read_csv(
'https://bcdanl.github.io/data/custdata_rev.csv')- We can load a CSV file directly from the web into R.
Getting to Know a data.frame
- The
$operator extracts a single column from adata.frameas avector.
dim()shows both the number of rows and columns.
nrow()andncol()give the row count and column count separately.
summary()gives a quick overview, whileskimr::skim()provides a more detailed, user-friendly summary of variables across all data types.
Observations in data.frame
- Rows in a
data.framerepresent individual units or entities for which data is collected.
- Examples:
- Student Information: Each row = one student
- Employee Information: Each row = one employee
- Daily S&P 500 Index Data: Each row = one trading day
- Household Survey Data: Each row = one household
- Student Information: Each row = one student
Variables in data.frame
- Columns in a
data.framerepresent attributes or characteristics measured across multiple observations.
- Examples:
- Student Data:
Name,Age,Grade,Major
- Employee Data:
EmployeeID,Name,Age,Department
- Customer Data:
CustomerID,Name,Age,Income,HousingType
- Student Data:
Note
- In a
data.frame, a variable is a column of data.
- In general programming, a variable is the name of an object.
Tidy data.frame
Variables, Observations, and Values

A
data.frameis tidy if it follows three rules:- Each variable has its own column.
- Each observation has its own row.
- Each value has its own cell.
- Each variable has its own column.
A tidy
data.framekeeps your data organized, making it easier to understand, analyze, and share in any data analysis.
Data Transformation with dplyr
Data Transformation with dplyr

dplyris a core tidyverse package for data manipulation — tasks like filtering, sorting, selecting, and renaming.
- Common
dplyrfunctions with data.frameDF:filter(DF, LOGICAL_CONDITIONS)
arrange(DF, VARIABLES)
distinct(DF, VARIABLES)
select(DF, VARIABLES)
rename(DF, NEW_NAME = CURRENT_NAME)
dplyrfunctions take a data.frame as the first argument.- They return a data.frame as output.
Making dplyr Code Flow with the Pipe Operator
- The pipe operator (
|>or%>%) makes code easier to read by connecting steps in order.
- How it works:
f(x, y)is the same asx |> f(y).
- Example with a data frame
DF:filter(DF, logical_condition)
- is the same as
DF |> filter(logical_condition)
- You can read the pipe as “then”:
- Example: Take the data, then filter it
Data Transformation Functions with the Pipe
- Common
dplyrfunctions with the pipe operator:DF |> filter(LOGICAL_CONDITIONS)
DF |> arrange(VARIABLES)
DF |> distinct(VARIABLES)
DF |> select(VARIABLES)
DF |> rename(NEW_NAME = CURRENT_NAME)
- Why this works so well:
dplyrfunctions usually take a data.frame as the first argument.
- They return a data.frame as output.
- This allows you to chain together multiple steps naturally:
- Example: Start with data, then filter, then arrange
\(\qquad\quad\;\;\)DF |> filter(...) |> arrange(...)
- Example: Start with data, then filter, then arrange
Using the Pipe in Posit Cloud
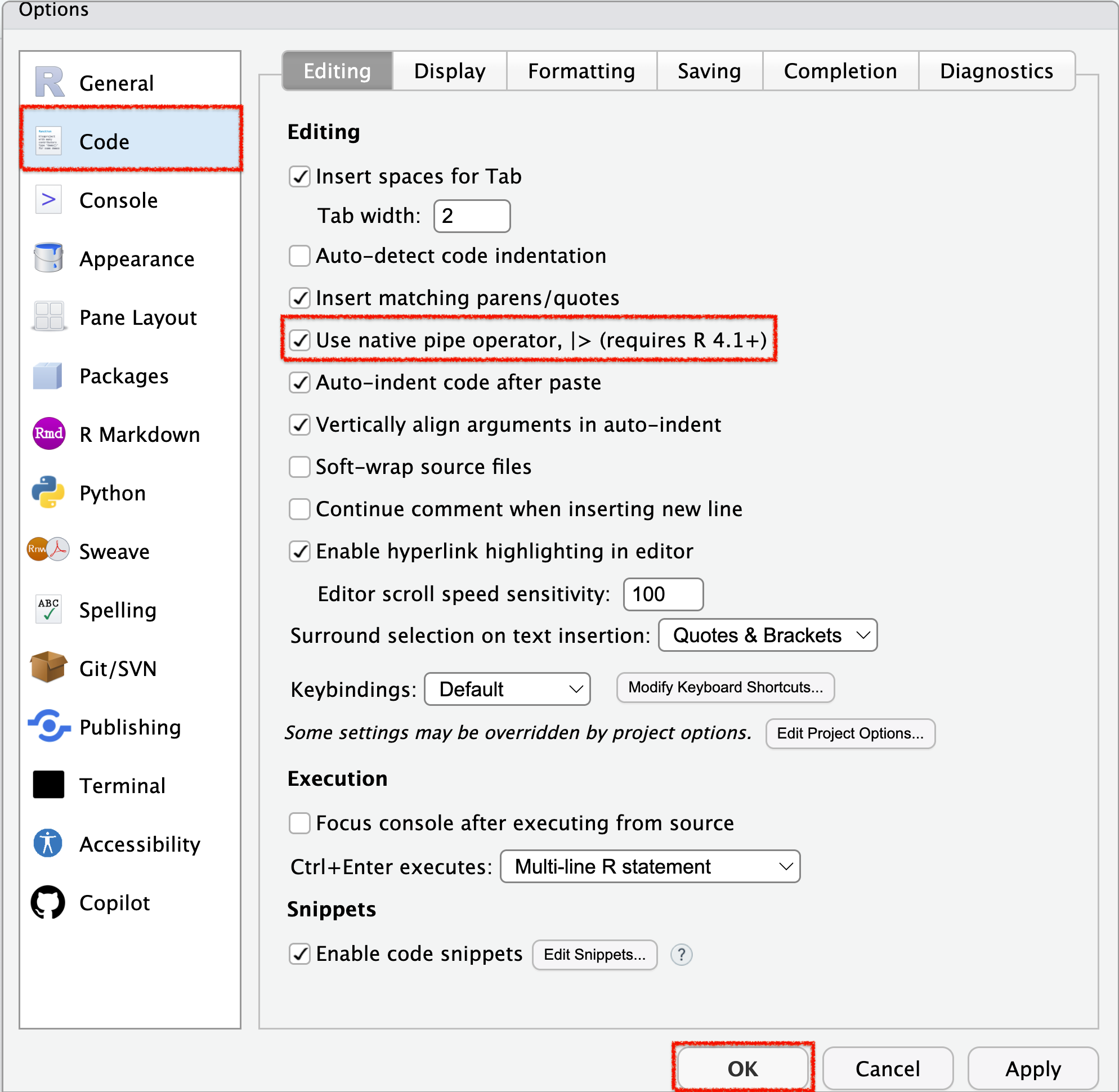
- To enable the native pipe operator (
|>) in RStudio:- Go to Tools > Global Options > Code (side menu).
- Under Pipe operator, choose Use native pipe operator (
|>).
- Go to Tools > Global Options > Code (side menu).
- Keyboard shortcut for inserting a pipe:
- Windows: Ctrl + Shift + M
- Mac: Cmd + Shift + M
- Windows: Ctrl + Shift + M
Filter observations with filter()
Filter Observations with filter()
install.packages("nycflights13") # Install once
library(nycflights13)
library(tidyverse)
flights <- nycflights13::flights
flights$month == 1 # A logical test returns TRUE or FALSE
class(flights$month == 12) filter()keeps only the observations that meet one or more logical conditions.- A logical condition evaluates to
TRUE(keep the observation) orFALSE(drop the observation).
- A logical condition evaluates to
Logical Conditions with Equality and Inequality
- Here, both
V1andV2are variables, and the comparisons are applied element-wise (vectorized).
- For logical conditions using inequalities, we focus on cases where
V1andV2areintegerornumeric.
Logical Conditions - Example 1
Logical Operators — AND, OR, NOT
- Here, both
xandyarelogicalconditions/variables.
- What logical operations (
&,|,!) do is combining logical variables/conditions, which returns alogicalvariable when executed.
Logical Operations
xandyare logical conditions.- If
xisTRUE, it highlights the left circle.
- If
yisTRUE, it highlights the right circle.
- If
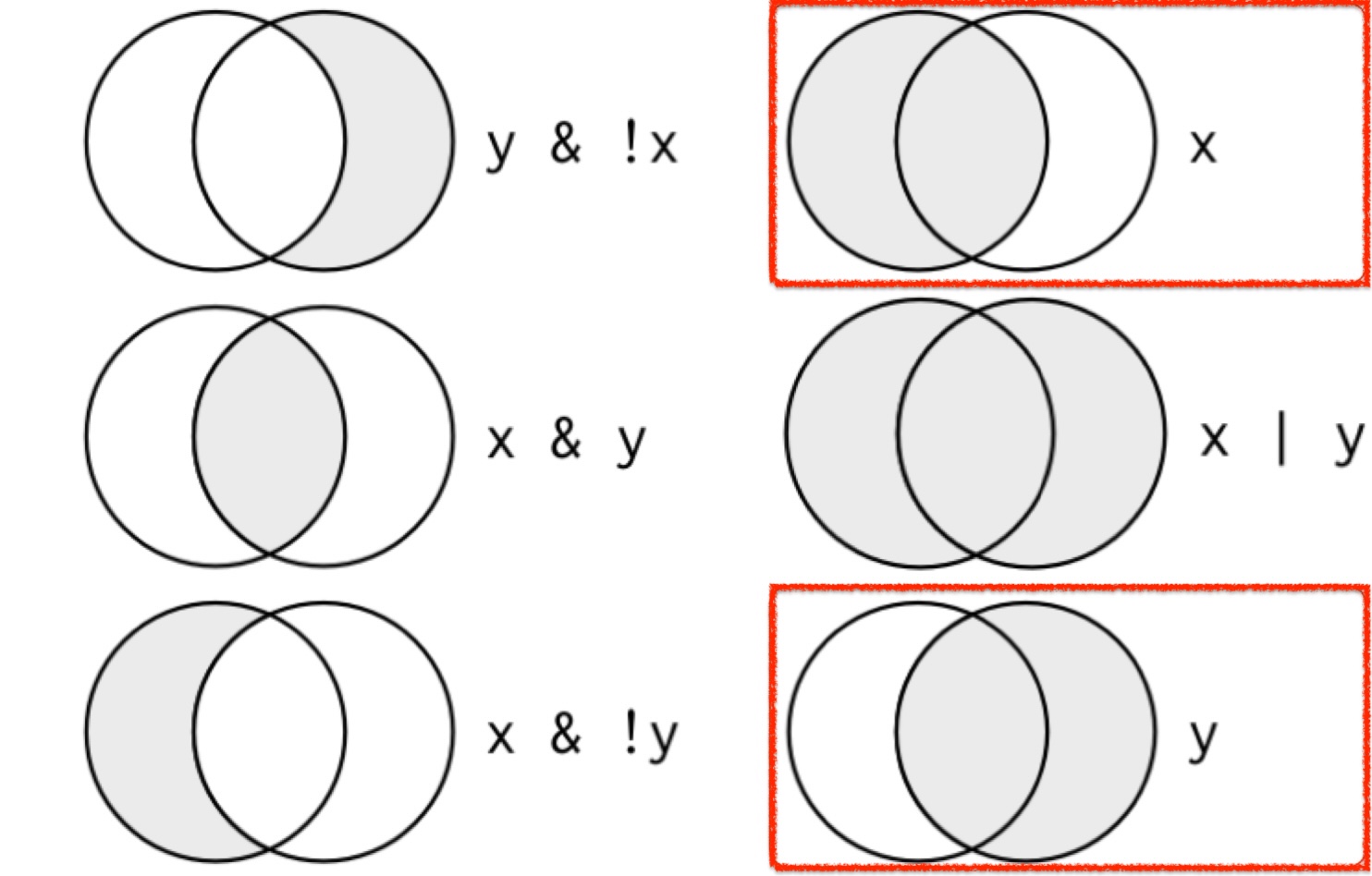
- The shaded regions show which parts each logical operator selects.
Logical Conditions - Example 2
- In
filter(), separating conditions with a comma is equivalent to combining them with the&operator.
Logical Conditions - Example 3
Logical Conditions - Example 4
Missing Values (NA)
NA(not available) represents a missing or unknown value in R.
- In most calculations, if one value is unknown, the result will also be unknown (
NA).
Comparing Missing Values
- Suppose
v1is Mary’s age (unknown) andv2is John’s age (unknown). - Can we say they are the same age?
- Since both values are missing, R cannot know — so the result is also
NA.
Checking for Missing Values with is.na()
- Use
is.na()to test whether a value is missing (NA).
- In
filter(), you can:- Use
is.na()to keep observations with missing values. - Use
!is.na()to remove observations with missing values.
- Use
Arrange Observations with arrange()
Arrange Observations with arrange()
arrange()sorts out observations.- When you provide multiple variables,
arrange()sorts by the first variable, and then uses the next variable(s) to break ties.
Descending Order with desc()
- Use
desc(VARIABLE)to sort in descending order.- For numeric variables, you can also use a leading minus sign.
Arrange Observations with arrange() - Example
- If we provide more than one variable name, each additional variable will be used to break ties in the values of preceding variables.
Find All Unique Observations with distinct()
Find All Unique Observations with distinct()
distinct()removes duplicate observations.- By default, it checks all variables to find unique observations.
Find All Unique Combinations of Selected Variables
- You can pass one or more variables to
distinct().- This returns unique combinations of just those variables (while ignoring the rest).
Select Variables with select()
Select Variables with select()
It’s not uncommon to get datasets with hundreds or thousands of variables.
select()allows us to narrow in on the variables we’re actually interested in.We can select variables by their names.
Remove Variables with select()
- With
select(-VARIABLES), we can remove variables.
Rename Variables with rename()
Rename Variables with rename()
rename()can be used to rename variables:DF |> rename(NEW_NAME = CURRENT_NAME)