Lecture 7
Linear Regression
February 17, 2025
Big Data and Machine Learning
Big Data and Machine Learning (ML)
- The term “Big Data” originated from computer scientists working with datasets too large to fit on a single machine.
- As aggregation evolved into analysis, Big Data became closely linked with statistics and machine learning (ML).
- Scalability of algorithms is crucial for handling large datasets.
- We will use ML to:
✅ Identify patterns in big data
✅ Make data-driven decisions
What does it mean to be “big”?
Big in both the number of observations (size
n) and in the number of variables (dimensionp).In these settings, we cannot:
- Look at each individual variable and make a decision.
- Choose among a small set of candidate models.
- Plot every variable to look for interactions or transformations.
Some ML tools are straight out of previous statistics classes (linear regression) and some are totally new (ensemble models, principal component analysis).
- All require a different approach when
nandpget really big.
- All require a different approach when
ML topics
- Regression: inference and prediction
- Regularization: cross-validation
- Principal Component Analysis: dimension reduction
- Tree-based models: decision trees, random forest, XGBoost
- Classfication: kNN
- Clustering: k-means, association rules
- Text Mining: sentiment analysis; topic models
Linear Regression Model
Models and Assumptions
Linear Model
- Linear regression assumes a linear relationship for \(Y = f(X_{1})\):
\[Y_{i} \,=\, \beta_{0} \,+\, \beta_{1} X_{1, i} \,+\, \epsilon_{i}\] for \(i \,=\, 1, 2, \dots, n\), where \(i\) is the \(i\)-th observation in data.
\(Y_i\) is the \(i\)-th value for the outcome/dependent/response/target variable \(Y\).
\(X_{1, i}\) is the \(i\)-th value for the explanatory/independent/predictor/input variable or feature \(X_{1}\).
Models and Assumptions
Beta coefficients
- Linear regression assumes a linear relationship for \(Y = f(X_{1})\):
\[Y_{i} \,=\, \beta_{0} \,+\, \beta_{1} X_{1, i} \,+\, \epsilon_{i}\] for \(i \,=\, 1, 2, \dots, n\), where \(i\) is the \(i\)-th observation in data.
\(\beta_0\) is an unknown true value of an intercept: average value for \(Y\) if \(X_{1} = 0\)
\(\beta_1\) is an unknown true value of a slope: increase in average value for \(Y\) for each one-unit increase in \(X_{1}\)
Models and Assumptions
Random Noises
- Linear regression assumes a linear relationship for \(Y = f(X_{1})\):
\[Y_{i} \,=\, \beta_{0} \,+\, \beta_{1} X_{1, i} \,+\, \epsilon_{i}\] for \(i \,=\, 1, 2, \dots, n\), where \(i\) is the \(i\)-th observation in data.
- \(\epsilon_i\) is a random noise, or a statistical error:
\[ \epsilon_i \sim N(0, \sigma^2) \]
- Errors have a mean value of 0 with constant variance \(\sigma^2\).
- Errors are uncorrelated with \(X_{1, i}\).
What Is Linear Regression Doing?
Best Fitting Line
Linear regression finds the beta estimates \(( \hat{\beta_{0}}, \hat{\beta_{1}} )\) such that:
– The linear function \(f(X_{1}) = \hat{\beta_{0}} + \hat{\beta_{1}}X_{1}\) is as near as possible to \(Y\) for all \((X_{1, i}\,,\, Y_{i})\) pairs in the data.
- It is the best fitting line, or the predicted outcome, \(\hat{Y_{\,}} = \hat{\beta_{0}} + \hat{\beta_{1}}X_{1}\).
What Is Linear Regression Doing?
Residual errors
- The estimated beta coefficients are chosen to minimize the sum of squares of the residual errors \((SSR)\): \[ \begin{align} SSR &\,=\, (\texttt{Residual_Error}_{1})^{2}\\ &\quad \,+\, (\texttt{Residual_Error}_{2})^{2}\\ &\quad\,+\, \cdots + (\texttt{Residual_Error}_{n})^{2}\\ \text{where}\qquad\qquad\qquad\qquad&\\ \texttt{Residual_Error}_{i} &\,=\, Y_{i} \,-\, \hat{Y_{i}},\\ \texttt{Predicted_Outcome}_{i}: \hat{Y_{i}} &\,=\, \hat{\beta_{0}} \,+\, \hat{\beta_{1}}X_{1, i} \end{align} \]
What Is Linear Regression Doing?
Hat Notation
We use the hat notation \((\,\hat{\texttt{ }_{\,}}\,)\) to distinguish true values and estimated/predicted values.
The value of true beta coefficient is denoted by \(\beta_{1}\).
The value of estimated beta coefficient is denoted by \(\hat{\beta_{1}}\).
The \(i\)-th value of true outcome variable is denoted by \(Y_{i}\).
The \(i\)-th value of predicted outcome variable is denoted by \(\hat{Y_{i}}\).
What Is Linear Regression Doing?
Relationship
1. Finding the relationship between \(X_{1}\) and \(Y\) \[\hat{\beta_{1}}\]: How is an increase in \(X_1\) by one unit associated with a change in \(Y\) on average? - Positive? Negative? Independent? - How strong?
Statistical Significance in Estimated Beta Coefficients
- What does it mean for a beta estimate \(\hat{\beta_{\,}}\) to be statistically significant at 5% level?
- It means that the null hypothesis \(H_{0}: \beta = 0\) is rejected for a given significance level 5%.
- “2 standard error rule” of thumb: The true value of \(\beta\) is 95% likely to be in the confidence interval \((\, \hat{\beta_{\,}} - 2 * \texttt{Std. Error}\;,\; \hat{\beta_{\,}} + 2 * \texttt{Std. Error} \,)\).
- The standard error tells us how uncertain our beta estimate is.
- We should look for the stars!
What Is Linear Regression Doing?
Prediction
2. Making a prediction on \(Y\): \[\hat{Y_{\,}}\] For unseen data point of \(X_1\), what is the predicted value of outcome, \(\hat{Y_{\,}}\)?
- E.g., For \(X_{1} = 2\), the predicted outcome is \(\hat{Y_{\,}} = \hat{\beta_{0}} + \hat{\beta_{1}} \times 2\).
- E.g., For \(X_{1} = 3\), the predicted outcome is \(\hat{Y_{\,}} = \hat{\beta_{0}} + \hat{\beta_{1}} \times 3\).
Linear Regression - Example
- Suppose we want to predict a property’s sales price based on the property size.
- In other words, for some house sale
i, we want to predictsale_price[i]based ongross_square_feet[i].
- In other words, for some house sale
- We also want to focus on the relationship between a property’s sales price and a property size.
- In other words, we estimate how an increase in
gross_square_feet[i]is associated withsale_price[i].
- In other words, we estimate how an increase in
Linear Relationship
Linear regression assumes that:
- The outcome
sale_price[i]is linearly related to the inputgross_square_feet[i]:
- The outcome
\[\texttt{sale_price[i]} \;=\quad \texttt{b0} \,+\, \texttt{b1*gross_square_feet[i]} \,+\, \texttt{e[i]}\] where e[i] is a statistical error term.
The Linear Relationship between sale_price and gross_square_feet
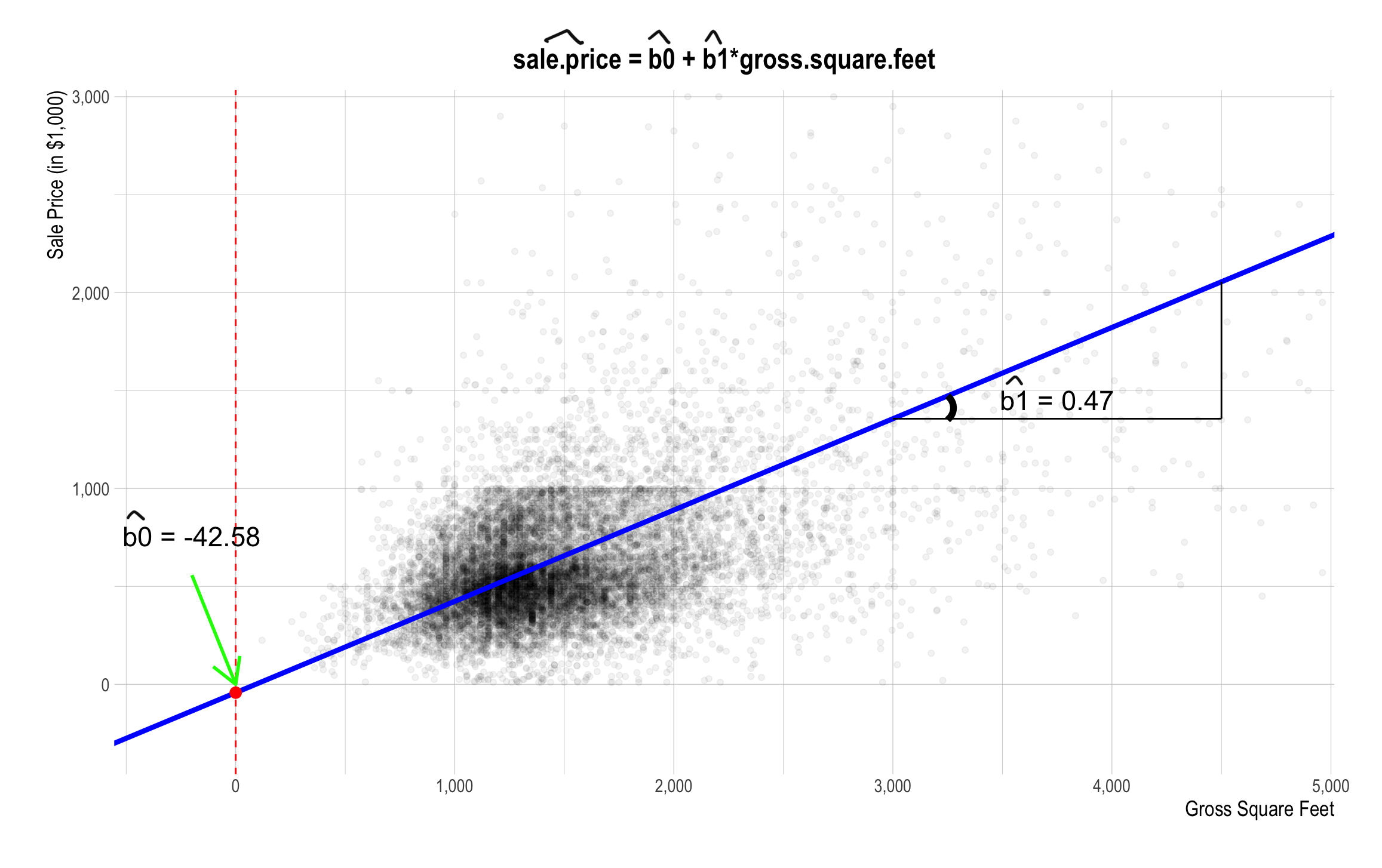
Best Fitting Line
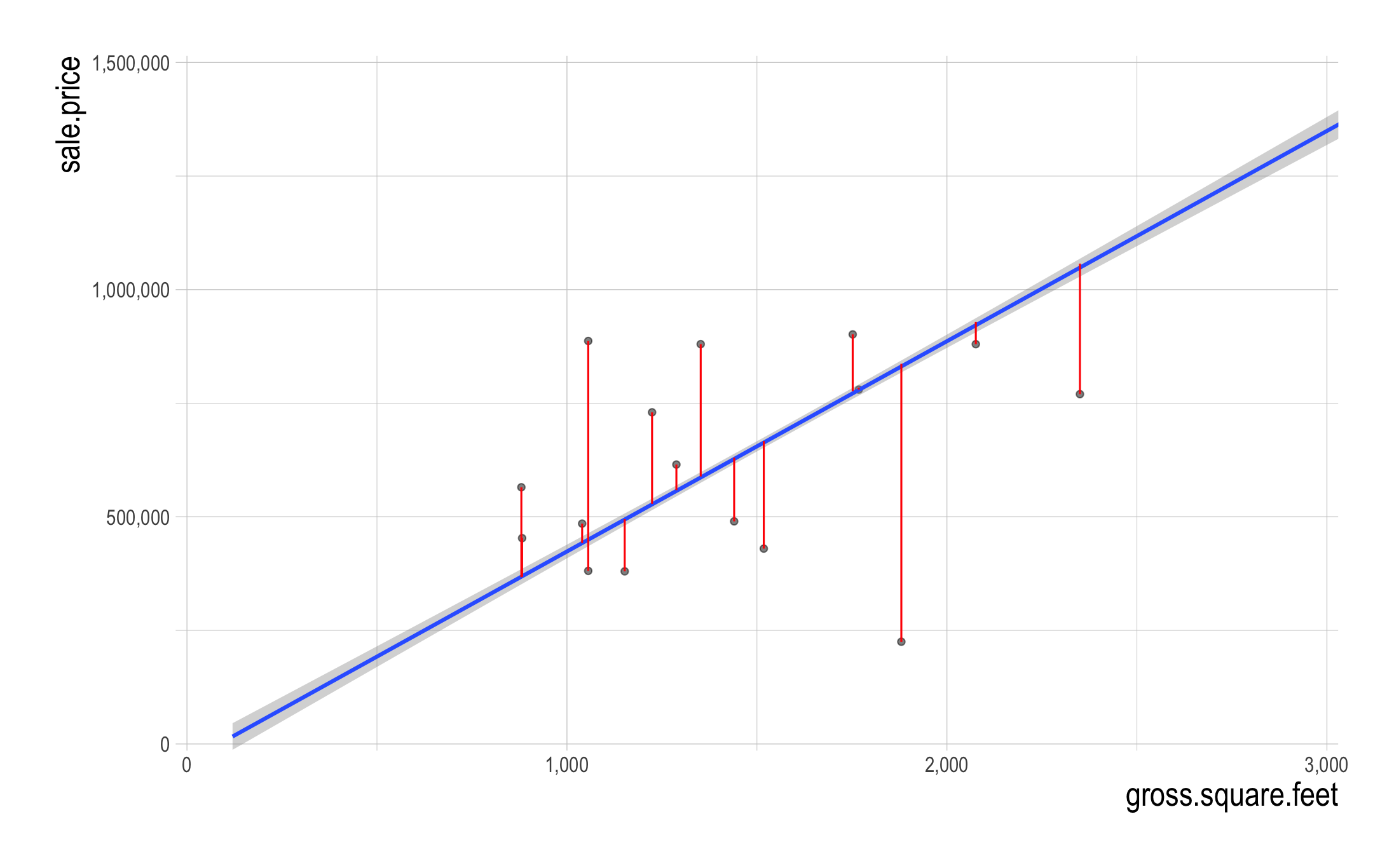
- What do the vertical lines visualize?
Model Evaluation
Mean squared error (MSE)
- One of the most common metrics used to measure the prediction accuracy of a linear regression model is MSE, which stands for mean squared error.
- \(MSE\) is \(SSR\) divided by \(n\) (the number of observations in the data that are used in making predictions).
\[ MSE = SSR / n \] \[ \begin{align} SSR &\,=\, (\texttt{Residual_Error}_{1})^{2}\\ &\quad \,+\, (\texttt{Residual_Error}_{2})^{2}\\ &\quad\,+\, \cdots + (\texttt{Residual_Error}_{n})^{2} \end{align} \]
- The lower MSE, the higher accuracy of the model.
- The root MSE (RMSE) is the square root of MSE.
Model Evaluation
Mean squared error (MSE)
- The root MSE (RMSE) represents the overall deviation of \(Y_{i}\) from the best fitting regression line.

Model Evaluation
R-squared
R-squared is a measure of how well the model “fits” the data, or its “goodness of fit.”
- R-squared can be thought of as what fraction of the
y’s variation is explained by the explanatory variables.
- R-squared can be thought of as what fraction of the
We want R-squared to be fairly large and R-squareds that are similar on testing and training.
Caution: R-squared will be higher for models with more explanatory variables, regardless of whether the additional explanatory variables actually improve the model or not.
Goals of Linear Regression
- The goals of linear regression here are to:
- Modeling for explanation: Find the relationship between
gross_square_feetandsale_priceby estimating a true value ofb1.
- The estimated value of
b1is denoted by \(\hat{\texttt{b1}}\).
- Modeling for prediction: Make a prediction on
sale_price[i]for new propertyi
- The predicted value of
sale_price[i]is denoted by \(\widehat{\texttt{sale_price}}\texttt{[i]}\), where
\[\widehat{\texttt{sale_price}}\texttt{[i]} \;=\quad \hat{\texttt{b0}} \,+\, \hat{\texttt{b1}}\texttt{*gross_square_feet[i]}\]
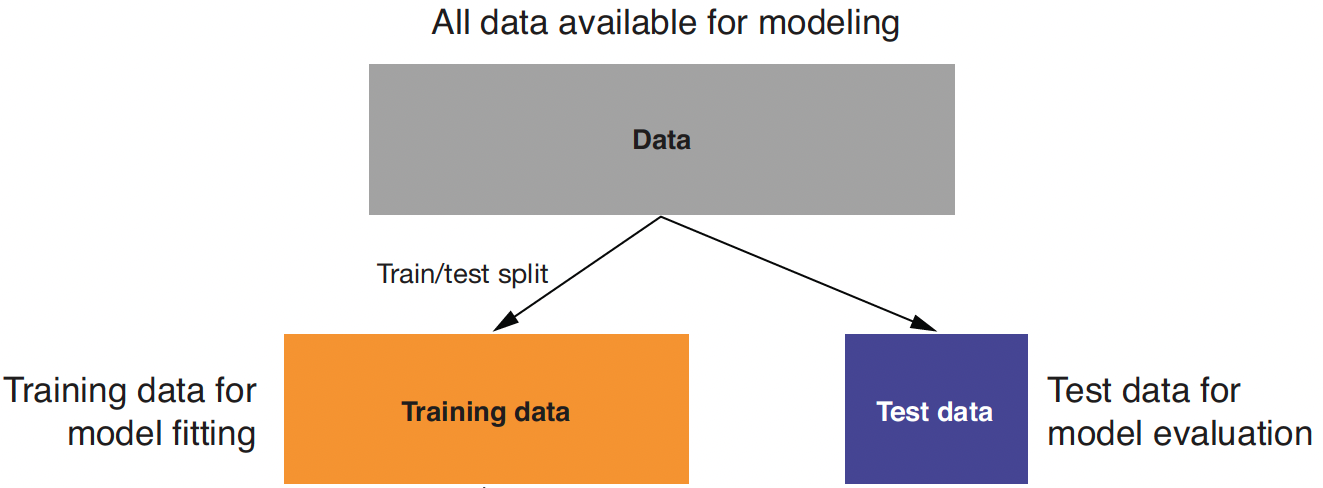
Training and Test Data
Training data: When we’re building a linear regression model, we need data to train the model.
Test data: We also need data to test whether the model works well on new data.
- So, we start with splitting a given data.frame into training and test data.frames when building a linear regression model.
Training and Test Data
Training vs. Test
- We use training data to train/fit the linear regression model.
- We then make a prediction using test data, which are unseen/new from the viewpoint of the trained linear regression model.
- In this way, we can test whether our model performs well in the real world, where unseen data points exist.
Training and Test Data
Overfitting
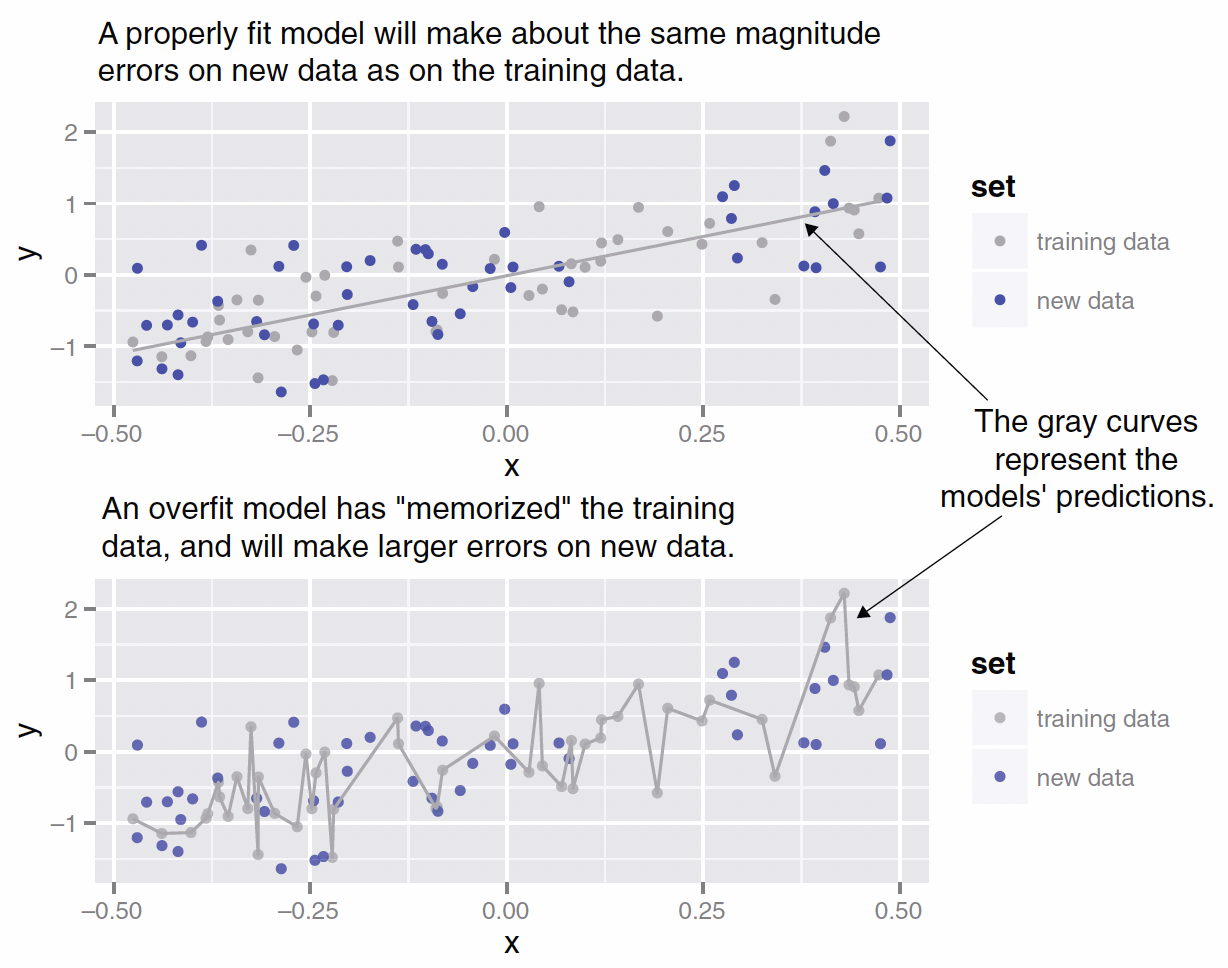
Training and Test Data
Model Construction and Evaluation
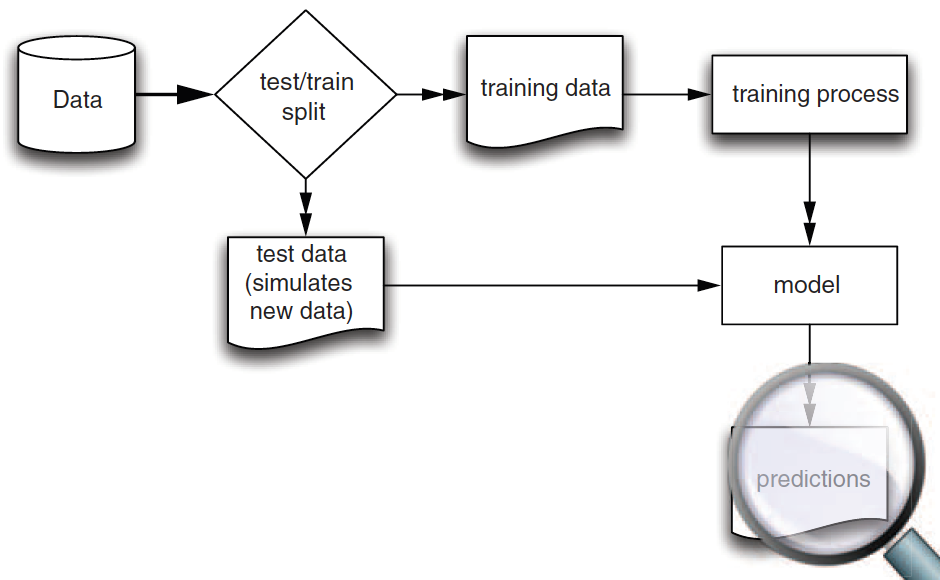
Splitting Data into Training and Testing Data
A Little Bit of Statistics for the Uniform Distribution
The probability density function for the uniform distribution looks like:
With the uniform distribution, any values of \(x\) between 0 and 1 is equally likely drawn.
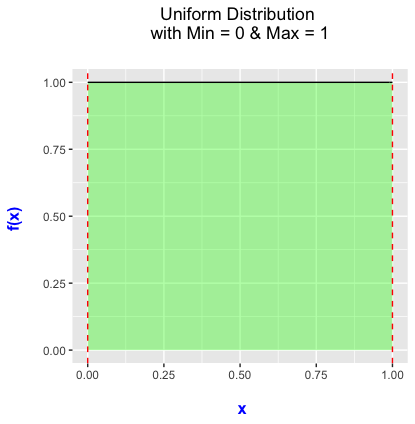
- We will use the uniform distribution when splitting data into training and testing data sets.
Splitting Data into Training and Testing Data
Randomization in the Sampling Process
- Why do we randomize when splitting given data into training and test data?
- Randomizing the sampling process ensures that the training and test data sets are representative for the population data.
- If the sample does not properly represent the entire population, the model result is biased toward the sample.
- Suppose the splitting process is not randomized, so that the observations with
sale_price > 10^6are in the training data and the observations withsale_price <= 10^6are in the test data.- What would be the model result then?
Linear Regression using PySpark
Example of Linear Regression using PySpark
We will use the data for residential property sales from September 2017 and August 2018 in NYC.
Each sales data recorded contains a number of interesting variables, but here we focus on the followings:
sale_price: a property’s sales price;gross_square_feet: a property’s size;age: a property’s age;borough_name: a borough where a property is located.
Use summary statistics and visualization to explore the data.
Splitting Data into Training and Testing Data
Step 1. Importing Modules and Reading DataFrames
import pandas as pd
import numpy as np
from tabulate import tabulate # for table summary
import scipy.stats as stats
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm # for lowess smoothing
from pyspark.sql import SparkSession
from pyspark.sql.functions import rand, col, pow, mean, avg, when, log, sqrt
from pyspark.ml.feature import VectorAssembler
from pyspark.ml.regression import LinearRegression
spark = SparkSession.builder.master("local[*]").getOrCreate()
# 1. Read CSV data from URL
df_pd = pd.read_csv('https://bcdanl.github.io/data/home_sales_nyc.csv')
sale_df = spark.createDataFrame(df_pd)
sale_df.show()Splitting Data into Training and Testing Data
Step 2. rand(seed = ANY_NUMBER)
# 2. Split data into training and testing sets by creating a random column "gp"
sale_df = sale_df.withColumn("gp", rand(seed=123)) # seed is set for replication
# Splits 60-40 into training and test sets
dtrain = sale_df.filter(col("gp") >= 0.4)
dtest = sale_df.filter(col("gp") < 0.4)
# Or simply,
dtrain, dtest = sale_df.randomSplit([0.6, 0.4], seed = 123)Building an ML DataFrame using VectorAssembler()
# Now assemble predictors using the renamed column
assembler1 = VectorAssembler(
inputCols=["gross_square_feet"],
outputCol="predictors")VectorAssembleris a transformer in PySpark’s ML library that is used to combine multiple columns into a single vector column.- Many ML algorithms in Spark require the predictors to be represented as a single vector.
VectorAssembleris often one of the first steps in a Spark ML pipeline.
Building an ML DataFrame using VectorAssembler()
dtrain1 = assembler1.transform(dtrain) # training data
dtest1 = assembler1.transform(dtest) # test data
dtrain1.select("predictors", "sale_price").show()
dtest1.select("predictors", "sale_price").show()VectorAssembler.transform()returns aDataFramewith a new column, specified inoutputColinVectorAssembler().
Building a Linear Regression Model using LinearRegression().fit()
# Fit linear regression model using the new label column "sale_price"
model1 = (
LinearRegression(
featuresCol = "predictors",
labelCol = "sale_price")
.fit(dtrain1)
)LinearRegression(featuresCol="predictors", labelCol="sale_price")- This creates an instance of the
LinearRegressionclass. - The features (independent variables) are in a column named “predictors”.
- The label (dependent variable) is in a column named “sale_price”.
- This creates an instance of the
.fit()trains (fits) the linear regression model using the training DataFramedtrain1.- This training process estimates the beta coefficients that best predict the label from the features.
Building a Linear Regression Model using LinearRegression().fit()
dtest1 = model1.transform(dtest1): Adds a new columnpredictiontodtest1DataFrame.- This new column contains the predicted outcome based on the trained
model1to predict an outcome for the test datasetdtest1.
- This new column contains the predicted outcome based on the trained
Summary of the Regression Result
Summary of the Regression Result
Example of a Regression Table
Summary of the Regression Result
Make the Summary Pretty
def regression_table(model, assembler):
"""
Creates a formatted regression table from a fitted LinearRegression model and its VectorAssembler.
If the model’s labelCol (retrieved using getLabelCol()) starts with "log", an extra column showing np.exp(coeff)
is added immediately after the beta estimate column for predictor rows. Additionally, np.exp() of the 95% CI
Lower and Upper bounds is also added unless the predictor's name includes "log_". The Intercept row does not
include exponentiated values.
When labelCol starts with "log", the columns are ordered as:
y: [label] | Beta | Exp(Beta) | Sig. | Std. Error | p-value | 95% CI Lower | Exp(95% CI Lower) | 95% CI Upper | Exp(95% CI Upper)
Otherwise, the columns are:
y: [label] | Beta | Sig. | Std. Error | p-value | 95% CI Lower | 95% CI Upper
Parameters:
model: A fitted LinearRegression model (with a .summary attribute and a labelCol).
assembler: The VectorAssembler used to assemble the features for the model.
Returns:
A formatted string containing the regression table.
"""
# Determine if we should display exponential values for coefficients.
is_log = model.getLabelCol().lower().startswith("log")
# Extract coefficients and standard errors as NumPy arrays.
coeffs = model.coefficients.toArray()
std_errors_all = np.array(model.summary.coefficientStandardErrors)
# Check if the intercept's standard error is included (one extra element).
if len(std_errors_all) == len(coeffs) + 1:
intercept_se = std_errors_all[0]
std_errors = std_errors_all[1:]
else:
intercept_se = None
std_errors = std_errors_all
# Use provided tValues and pValues.
df = model.summary.numInstances - len(coeffs) - 1
t_critical = stats.t.ppf(0.975, df)
p_values = model.summary.pValues
# Helper: significance stars.
def significance_stars(p):
if p < 0.01:
return "***"
elif p < 0.05:
return "**"
elif p < 0.1:
return "*"
else:
return ""
# Build table rows for each feature.
table = []
for feature, beta, se, p in zip(assembler.getInputCols(), coeffs, std_errors, p_values):
ci_lower = beta - t_critical * se
ci_upper = beta + t_critical * se
# Check if predictor contains "log_" to determine if exponentiation should be applied
apply_exp = is_log and "log_" not in feature.lower()
exp_beta = np.exp(beta) if apply_exp else ""
exp_ci_lower = np.exp(ci_lower) if apply_exp else ""
exp_ci_upper = np.exp(ci_upper) if apply_exp else ""
if is_log:
table.append([
feature, # Predictor name
beta, # Beta estimate
exp_beta, # Exponential of beta (or blank)
significance_stars(p),
se,
p,
ci_lower,
exp_ci_lower, # Exponential of 95% CI lower bound
ci_upper,
exp_ci_upper # Exponential of 95% CI upper bound
])
else:
table.append([
feature,
beta,
significance_stars(p),
se,
p,
ci_lower,
ci_upper
])
# Process intercept.
if intercept_se is not None:
intercept_p = model.summary.pValues[0] if model.summary.pValues is not None else None
intercept_sig = significance_stars(intercept_p)
ci_intercept_lower = model.intercept - t_critical * intercept_se
ci_intercept_upper = model.intercept + t_critical * intercept_se
else:
intercept_sig = ""
ci_intercept_lower = ""
ci_intercept_upper = ""
intercept_se = ""
if is_log:
table.append([
"Intercept",
model.intercept,
"", # Removed np.exp(model.intercept)
intercept_sig,
intercept_se,
"",
ci_intercept_lower,
"",
ci_intercept_upper,
""
])
else:
table.append([
"Intercept",
model.intercept,
intercept_sig,
intercept_se,
"",
ci_intercept_lower,
ci_intercept_upper
])
# Append overall model metrics.
if is_log:
table.append(["Observations", model.summary.numInstances, "", "", "", "", "", "", "", ""])
table.append(["R²", model.summary.r2, "", "", "", "", "", "", "", ""])
table.append(["RMSE", model.summary.rootMeanSquaredError, "", "", "", "", "", "", "", ""])
else:
table.append(["Observations", model.summary.numInstances, "", "", "", "", ""])
table.append(["R²", model.summary.r2, "", "", "", "", ""])
table.append(["RMSE", model.summary.rootMeanSquaredError, "", "", "", "", ""])
# Format the table rows.
formatted_table = []
for row in table:
formatted_row = []
for i, item in enumerate(row):
# Format Observations as integer with commas.
if row[0] == "Observations" and i == 1 and isinstance(item, (int, float, np.floating)) and item != "":
formatted_row.append(f"{int(item):,}")
elif isinstance(item, (int, float, np.floating)) and item != "":
if is_log:
# When is_log, the columns are:
# 0: Metric, 1: Beta, 2: Exp(Beta), 3: Sig, 4: Std. Error, 5: p-value,
# 6: 95% CI Lower, 7: Exp(95% CI Lower), 8: 95% CI Upper, 9: Exp(95% CI Upper).
if i in [1, 2, 4, 6, 7, 8, 9]:
formatted_row.append(f"{item:,.3f}")
elif i == 5:
formatted_row.append(f"{item:.3f}")
else:
formatted_row.append(f"{item:.3f}")
else:
# When not is_log, the columns are:
# 0: Metric, 1: Beta, 2: Sig, 3: Std. Error, 4: p-value, 5: 95% CI Lower, 6: 95% CI Upper.
if i in [1, 3, 5, 6]:
formatted_row.append(f"{item:,.3f}")
elif i == 4:
formatted_row.append(f"{item:.3f}")
else:
formatted_row.append(f"{item:.3f}")
else:
formatted_row.append(item)
formatted_table.append(formatted_row)
# Set header and column alignment based on whether label starts with "log"
if is_log:
headers = [
f"y: {model.getLabelCol()}",
"Beta", "Exp(Beta)", "Sig.", "Std. Error", "p-value",
"95% CI Lower", "Exp(95% CI Lower)", "95% CI Upper", "Exp(95% CI Upper)"
]
colalign = ("left", "right", "right", "center", "right", "right", "right", "right", "right", "right")
else:
headers = [f"y: {model.getLabelCol()}", "Beta", "Sig.", "Std. Error", "p-value", "95% CI Lower", "95% CI Upper"]
colalign = ("left", "right", "center", "right", "right", "right", "right")
table_str = tabulate(
formatted_table,
headers=headers,
tablefmt="pretty",
colalign=colalign
)
# Insert a dashed line after the Intercept row.
lines = table_str.split("\n")
dash_line = '-' * len(lines[0])
for i, line in enumerate(lines):
if "Intercept" in line and not line.strip().startswith('+'):
lines.insert(i+1, dash_line)
break
return "\n".join(lines)
# Example usage:
# print(regression_table(model_1, assembler_1))Summary of the Regression Result
Make the Summary Pretty
Linear Regression Model with Multiple Predictors
Multiple Regression
- What if the regression were missing something?
- Maybe prices are not just about size, but maybe there are certain parts of NYC that are categorically more expensive than other parts of NYC.
- Maybe Manhattan is just more expensive than Bronx.
- Maybe apartments are different than non-apartments.
- Maybe old houses are different than new houses.
- It is often helpful to bring in multiple explanatory variables—a Multivariate Regression.
Models and Assumptions
- Linear regression assumes a linear relationship for \(Y = f(X_{1}, X_{2})\):
\[Y_{i} \,=\, \beta_{0} \,+\, \beta_{1} X_{1, i} \,+\,\beta_{2} X_{2, i} \,+\, \epsilon_{i}\] for \(i \,=\, 1, 2, \dots, n\), where \(i\) is the \(i\)-th observation in data.
\(\beta_0\) is an unknown true value of an intercept: average value for \(Y\) if \(X_{1} = 0\) and \(X_{2} = 0\)
\(\beta_1\) is an unknown true value of a slope: increase in average value for \(Y\) for each one-unit increase in \(X_{1}\)
\(\beta_2\) is an unknown true value of a slope: increase in average value for \(Y\) for each one-unit increase in \(X_{2}\)
Models and Assumptions
Random Noises
- Linear regression assumes a linear relationship for \(Y = f(X_{1}, X_{2})\):
\[Y_{i} \,=\, \beta_{0} \,+\, \beta_{1} X_{1, i}\,+\, \beta_{1} X_{2, i} \,+\, \epsilon_{i}\] for \(i \,=\, 1, 2, \dots, n\), where \(i\) is the \(i\)-th observation in data.
- \(\epsilon_i\) is a random noise, or a statistical error:
\[ \epsilon_i \sim N(0, \sigma^2) \]
- Errors have a mean value of 0 with constant variance \(\sigma^2\).
- Errors are uncorrelated with \(X_{1, i}\) and with \(X_{2, i}\).
Models and Assumptions
Best Fitting Plane
Linear regression finds the beta estimates \(( \hat{\beta_{0}}, \hat{\beta_{1}}, \hat{\beta_{2}} )\) such that:
– The linear function \(f(X_{1}, X_{2}) = \hat{\beta_{0}} + \hat{\beta_{1}}X_{1} + \hat{\beta_{2}}X_{2}\) is as near as possible to \(Y\) for all \((X_{1, i}\,,\,X_{2, i}\,,\, Y_{i})\) pairs in the data.
- It is the best fitting plane, or the predicted outcome, \(\hat{Y_{\,}} = \hat{\beta_{0}} + \hat{\beta_{1}}X_{1} + \hat{\beta_{2}}X_{2}\)
Models and Assumptions
Residual Errors
- The estimated beta coefficients are chosen to minimize the sum of squares of the residual errors \((SSR)\): \[ \begin{align} SSR &\,=\, (\texttt{Residual_Error}_{1})^{2}\\ &\quad \,+\, (\texttt{Residual_Error}_{2})^{2}\\ &\quad\,+\, \cdots + (\texttt{Residual_Error}_{n})^{2}\\ \text{where}\qquad\qquad\qquad\qquad&\\ \texttt{Residual_Error}_{i} &\,=\, Y_{i} \,-\, \hat{Y_{i}},\\ \texttt{Predicted_Outcome}_{i}: \hat{Y_{i}} &\,=\, \hat{\beta_{0}} \,+\, \hat{\beta_{1}}X_{1, i} \,+\, \hat{\beta_{2}}X_{2, i} \end{align} \]
Multiple Regression - Best Fitting Plane
- All else being equal, an increase in
gross_square_feetby one unit is associated with an increase insale_priceby \(\hat{\beta_{1}}\).
Multiple Regression using PySpark
- Let’s add a new predictor,
age, to the model.
assembler2 = VectorAssembler(
inputCols=["gross_square_feet", "age"],
outputCol="predictors")
dtrain2 = assembler2.transform(dtrain)
dtest2 = assembler2.transform(dtest)
model2 = LinearRegression(
featuresCol="predictors",
labelCol="sale_price")
.fit(dtrain2)
dtest2 = model2.transform(dtest2)
print(regression_table(model2, assembler2))Dummy Variables in Linear Regression
Motivation: Treating Categorical Variables in Linear Regression
Linear regression models require numerical predictors, but many variables are categorical.
- e.g., Consider the two equivalent houses, except for its location—one in Manhattan and the other in Bronx.
The Approach:
Convert categorical variables into numerical format using dummy variables.Why Do This?
- Allows the model to compare different categories.
- Each dummy variable indicates the presence (1) or absence (0) of a category.
What are Dummy Variables?
- Definition: Binary indicators (0 or 1) representing categorical data.
- Purpose: Transform qualitative data into a quantitative form for regression analysis.
Example:
\[ D_i = \begin{cases} 1, & \text{if the observation belongs to the category} \\ 0, & \text{otherwise} \end{cases} \]
Dummy Variables in Regression Models
Consider a regression model including a dummy variable:
\[ y_i = \beta_0 + \beta_1 x_i + \beta_2 D_i + \epsilon_i \]
- \(x_i\): A continuous predictor.
- \(D_i\): Dummy variable (e.g., political party affiliation, type of car).
Interpretation:
\(\beta_2\) captures the difference in the response \(y\) when the category is present (i.e., \(D_i=1\)) versus absent.
The Dummy Variable Trap
Problem: Including a dummy for every category introduces redundancy.
For a categorical variable with \(k\) levels:
- If you include all \(k\) dummy variables in the model, their values always sum to 1:
\[ D_{1i} + D_{2i} + \cdots + D_{ki} = 1 \quad \text{(for each observation)} \]
This is problematic, because one dummy is completely predictable from the others.
The intercept already captures the constant part (1), making one of the dummy variables redundant.
Avoiding the Dummy Variable Trap
Solution: Drop one dummy (often called the reference category)
The reference category is represented by a combination of \(\texttt{borough}\) variables.
- Dummy for the reference category is 1 if all the rest of the dummies is 0.
- Dummy for the reference category is 0 otherwise.
Proper model:
\[ y_i = \beta_0 + \beta_1 D_{1, i} + \beta_2 D_{2, i} + \cdots + \beta_{k-1} D_{(k-1), i} + \epsilon_i \]
- Interpretation:
- Each \(\beta_j\) (for \(j=1,2,\ldots,k-1\)) represents the difference from the reference category.
Dummy Variable Regression using PySpark
- The UDF,
add_dummy_variables(var_name, reference_level)convert a categorical variable into its dummy variables:var_name: a string of categorical variable namereference_level: index position of alphabetically sorted categories- This function requires
dtrainanddtestDataFrames.
def add_dummy_variables(var_name, reference_level, category_order=None):
"""
Creates dummy variables for the specified column in the global DataFrames dtrain and dtest.
Allows manual setting of category order.
Parameters:
var_name (str): The name of the categorical column (e.g., "borough_name").
reference_level (int): Index of the category to be used as the reference (dummy omitted).
category_order (list, optional): List of categories in the desired order. If None, categories are sorted.
Returns:
dummy_cols (list): List of dummy column names excluding the reference category.
ref_category (str): The category chosen as the reference.
"""
global dtrain, dtest
# Get distinct categories from the training set.
categories = dtrain.select(var_name).distinct().rdd.flatMap(lambda x: x).collect()
# Convert booleans to strings if present.
categories = [str(c) if isinstance(c, bool) else c for c in categories]
# Use manual category order if provided; otherwise, sort categories.
if category_order:
# Ensure all categories are present in the user-defined order
missing = set(categories) - set(category_order)
if missing:
raise ValueError(f"These categories are missing from your custom order: {missing}")
categories = category_order
else:
categories = sorted(categories)
# Validate reference_level
if reference_level < 0 or reference_level >= len(categories):
raise ValueError(f"reference_level must be between 0 and {len(categories) - 1}")
# Define the reference category
ref_category = categories[reference_level]
print("Reference category (dummy omitted):", ref_category)
# Create dummy variables for all categories
for cat in categories:
dummy_col_name = var_name + "_" + str(cat).replace(" ", "_")
dtrain = dtrain.withColumn(dummy_col_name, when(col(var_name) == cat, 1).otherwise(0))
dtest = dtest.withColumn(dummy_col_name, when(col(var_name) == cat, 1).otherwise(0))
# List of dummy columns, excluding the reference category
dummy_cols = [var_name + "_" + str(cat).replace(" ", "_") for cat in categories if cat != ref_category]
return dummy_cols, ref_category
# Example usage without category_order:
# dummy_cols_year, ref_category_year = add_dummy_variables('year', 0)
# Example usage with category_order:
# custom_order_wkday = ['sunday', 'monday', 'tuesday', 'wednesday', 'thursday', 'friday', 'saturday']
# dummy_cols_wkday, ref_category_wkday = add_dummy_variables('wkday', reference_level=0, category_order = custom_order_wkday)Dummy Variable Regression using PySpark
- Let’s check what categories are in “borough_name” and how many categories are:
- Let’s convert the borough_name variable into its dummies using the UDF,
add_dummy_variables(var_name, reference_level):
# 0: Bronx, 1: Brooklyn, 2: Manhattan, 3: Queens, 4: Staten Island
dummy_cols, ref_category = add_dummy_variables("borough_name", 2)
# To see dummy variables
dtrain.select(['borough_name'] + dummy_cols).show()
dtrain.select(['borough_name'] + dummy_cols).filter(col('borough_name') == "Manhattan").show()Dummy Variable Regression using PySpark
- Now we’re ready to run a linear regression with dummy variables:
# Define the list of predictor columns:
# Two numeric predictors plus the dummy variables (excluding the reference dummy)
conti_cols = ["gross_square_feet", "age"]
assembler_predictors = conti_cols + dummy_cols
assembler3 = VectorAssembler(
inputCols = assembler_predictors,
outputCol = "predictors"
)
dtrain3 = assembler3.transform(dtrain)
dtest3 = assembler3.transform(dtest)
model3 = LinearRegression(featuresCol="predictors", labelCol="sale_price").fit(dtrain3)
dtest3 = model3.transform(dtest3)
# For model3 and assembler3:
print(regression_table(model3, assembler3))Residual Plots
Residuals
- Model equation: \(Y_{i} \,=\, \beta_{0} \,+\, \beta_{1}X_{1,i} \,+\, \beta_{2}X_{2,i}\)
- \(\epsilon_i\) is a random noise, or a statistical error:
\[ \epsilon_i \sim N(0, \sigma^2) \] - Errors have a mean value of 0 with constant variance \(\sigma^2\).
- Errors are uncorrelated with \(X_{1,i}\) and with \(X_{2, i}\).
Residuals
If we re-arrange the simple regression equation, \[\begin{align} {\epsilon}_{i} \,=\, Y_{i} \,-\, (\, {\beta}_{0} \,+\, {\beta}_{1}X_{1,i} \,). \end{align}\]
\(\texttt{residual_error}_{i}\) can be thought of as the expected value of \(\epsilon_{i}\), denoted by \(\hat{\epsilon_{i}}\). \[\begin{align} \hat{\epsilon_{i}} \,=\, Y_{i} \,-\, (\, \hat{\beta_{0}} \,+\, \hat{\beta_{1}}X_{1,i} \,) \end{align}\]
Residual Plots
Residual plot is a scatterplot of fitted values and residuals.
- A variable of fitted values on x-axis
- A variable of residuals on y-axis
A residual plot can be used to diagnose the quality of model results.
Because we assume that \(\epsilon_{i}\) have a mean value of 0 with constant variance \(\sigma^2\), a well-behaved residual plot should bounce randomly and form a cloud roughly at the level of zero residual, the perfect prediction line.
From the residual plot, we should ask the following the two questions ourselves:
- On average, are the predictions correct?
- Are there systematic errors?
Residual Plots
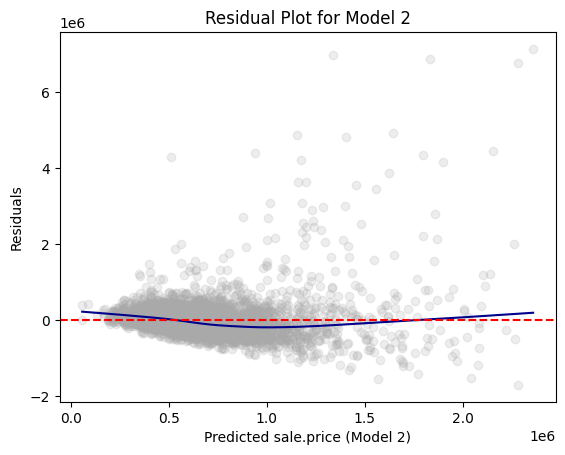
- On average, are the predictions correct?
- Are there systematic errors?
Residual Plots
Unbiasedness and Homoskedasticity
- We would like have a residual plot to be
- Unbiased: have an average value of zero in any thin vertical strip;
- Homoskedastic, which means “same stretch”: it is ideal to have the same spread of the residuals in any thin vertical strip.
- When the variance of residuals changes across predicted values (e.g., residuals get larger as predicted values increase), the model suffers from heteroscedasticity.
Residual Plots
Examples
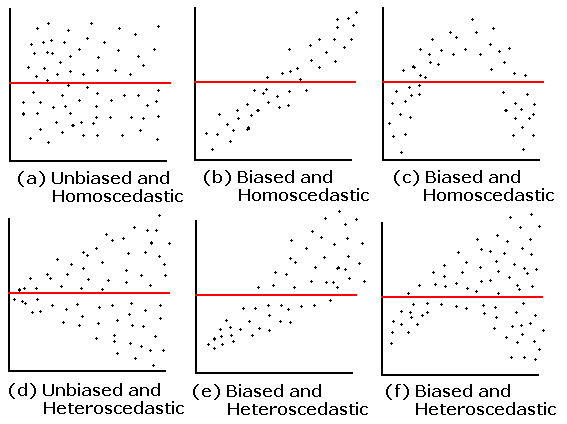
Residual Plots
What happens if biased?
- The model consistently overpredicts or underpredicts for certain values.
- Indicates that the model might be misspecified—perhaps missing important variables or using an incorrect functional form.
- Leads to biased parameter estimates, meaning the coefficients are systematically off, reducing the validity of predictions and inferences.
Residual Plots
What happens if heteroscedasticity is present?
- Consequences of heteroscedasticity:
- 📉 Inefficient coefficient estimates: Estimates remain unbiased but are no longer efficient (i.e., they don’t have the smallest variance).
- ❌ Biased standard errors: Leads to unreliable p-values and confidence intervals, potentially resulting in invalid hypothesis tests.
- ⚠️ Misleading inferences: Predictors may appear statistically significant or insignificant incorrectly.
- 🎯 Poor predictive performance: The model might perform poorly on future data, especially if the residual variance grows with higher predicted values.
Residual Plots in Python
- PySpark itself does not have built-in visualization capabilities.
- Firstly, we convert PySpark
DataFrameinto PandasDataFrameby using.toPandas()
- Firstly, we convert PySpark
Residual Plots in Python
- We then use
matplotlib.pyplotto draw a residual plot:
import matplotlib.pyplot as plt
import statsmodels.api as sm # for lowess smoothing
plt.scatter(rdf["prediction"], rdf["residual"], alpha=0.2, color="darkgray")
# Use lowess smoothing for the trend line
smoothed = sm.nonparametric.lowess(rdf["residual"], rdf["prediction"])
plt.plot(smoothed[:, 0], smoothed[:, 1], color="darkblue")
# Perfect prediction line
# A horizontal line at residual = 0
plt.axhline(y=0, color="red", linestyle="--")
# Labeling
plt.xlabel("Predicted sale.price (Model 2)")
plt.ylabel("Residuals")
plt.title("Residual Plot for Model 2")
plt.show()Hypothesis Testing on Beta Coefficient
Hypothesis Testing on Beta Coefficient
To determine whether an independent variable has a statistically significant effect on the dependent variable in a linear regression model.
Consider the following linear regression model: \[ y_{i} = \beta_0 + \beta_1 x_{1,i} + \beta_2 x_{2,i} + \dots + \beta_k x_{k,i} + \epsilon_{i} \]
\(y\): Outcome
\(x_1, x_2, \dots, x_k\): Predictors
\(\beta_0\): Intercept
\(\beta_1, \beta_2, \dots, \beta_k\): Coefficients
\(\epsilon_{i}\): Error term
Hypotheses and Test Statistic
- We test whether a specific coefficient \(\beta_j\) significantly differs from zero:
- Null Hypothesis (\(H_0\)): \(\beta_j = 0\) (No relationship)
- Alternative Hypothesis (\(H_A\)): \(\beta_j \neq 0\) (Significant relationship)
- Null Hypothesis (\(H_0\)): \(\beta_j = 0\) (No relationship)
- The t-statistic is used to test each coefficient:
\[ t = \frac{\hat{\beta_j} - 0}{SE(\hat{\beta_j})} \]
- \(\hat{\beta_j}\): Estimated coefficient
- \(SE(\hat{\beta_j})\): Standard error of the estimate
Hypothesis Testing - Decision Rule and Interpretation
- Calculate the p-value based on the t-distribution with \(n - k - 1\) degrees of freedom.
- Compare p-value with significance level \(\alpha\) (e.g., 0.05):
- If \(p \leq \alpha\): Reject \(H_0\) (Significant)
- If \(p > \alpha\): Fail to reject \(H_0\) (Not significant)
- In our course, stars in a regression table mean a significance level:
*(10%);**(5%);***(1%)
- Reject \(H_0\): There is sufficient evidence to suggest a statistically significant relationship between \(x_j\) and \(y\).
- Fail to reject \(H_0\): No statistically significant evidence of a relationship between \(x_j\) and \(y\).
Interpreting Beta Estimates
Interpreting Estimated Beta Coefficients
Example
The model equation is \[\begin{align}
\texttt{sale_price[i]} \;=\;\, &\texttt{b0} \,+\,\\ &\texttt{b1*gross_square_feet[i]} \,+\,\texttt{b2*age[i]}\,+\,\\ &\texttt{b3*Bronx[i]} \,+\,\texttt{b4*Brooklyn[i]} \,+\,\\&\texttt{b5*Queens[i]} \,+\,\texttt{b6*Staten Island[i]}\,+\,\\ &\texttt{e[i]}
\end{align}\] - The reference level of borough_name variables is Manhattan.
Interpreting Estimated Beta Coefficients
1. gross_square_feet
- Consider the predicted sales prices of the two houses,
AandB.- Both
AandBare inBronxand with the sameage. gross_square_feetof houseAis 2001, while that of houseBis 2000.
- Both
- All else being equal, an increase in
gross_square_feetby one unit is associated with an increase insale_priceby \(\hat{\beta_{1}}\).- Why?
Interpreting Estimated Beta Coefficients
1. gross_square_feet
\[ \begin{align}\widehat{\texttt{sale_price[A]}} \;=\quad& \hat{\texttt{b0}} \,+\, \hat{\texttt{b1}}\texttt{*gross_square_feet[A]} \,+\, \hat{\texttt{b2}}\texttt{*age[A]} \,+\,\\ &\hat{\texttt{b3}}\texttt{*Bronx[A]}\,+\,\hat{\texttt{b4}}\texttt{*Brooklyn[A]} \,+\,\\ &\hat{\texttt{b5}}\texttt{*Queens[A]}\,+\, \hat{\texttt{b6}}\texttt{*Staten Island[A]}\\ \widehat{\texttt{sale_price[B]}} \;=\quad& \hat{\texttt{b0}} \,+\, \hat{\texttt{b1}}\texttt{*gross_square_feet[B]} \,+\, \hat{\texttt{b2}}\texttt{*age[B]}\,+\,\\ &\hat{\texttt{b3}}\texttt{*Bronx[B]}\,+\, \hat{\texttt{b4}}\texttt{*Brooklyn[B]} \,+\,\\ &\hat{\texttt{b5}}\texttt{*Queens[B]}\,+\, \hat{\texttt{b6}}\texttt{*Staten Island[B]} \end{align} \]
\[ \begin{align}\Leftrightarrow\qquad&\widehat{\texttt{sale_price[A]}} \,-\, \widehat{\texttt{sale_price[B]}}\qquad \\ \;=\quad &\hat{\texttt{b1}}\texttt{*}(\texttt{gross_square_feet[A]} - \texttt{gross_square_feet[B]})\\ \;=\quad &\hat{\texttt{b1}}\texttt{*}\texttt{(2001 - 2000)} \,=\, \hat{\texttt{b1}}\qquad\qquad\quad\;\; \end{align} \]
Interpreting Estimated Beta Coefficients
2. borough_nameBronx
Consider the predicted sales prices of the two houses,
AandC.- Both
AandCare with the sameageand the samegross_square_feet. Ais inBronx, andCis inManhattan.
- Both
All else being equal, an increase in
borough_nameBronxby one unit is associated with an increase insale_pricebyb3.Equivalently, all else being equal, being in
Bronxrelative to being a inManhattanis associated with a decrease insale_priceby|b3|.- Why?
Interpreting Estimated Beta Coefficients
2. borough_nameBronx
\[ \begin{align}\widehat{\texttt{sale_price[A]}} \;=\quad& \hat{\texttt{b0}} \,+\, \hat{\texttt{b1}}\texttt{*gross_square_feet[A]} \,+\, \hat{\texttt{b2}}\texttt{*age[A]} \,+\,\\ &\hat{\texttt{b3}}\texttt{*Bronx[A]}\,+\, \hat{\texttt{b4}}\texttt{*Brooklyn[A]} \,+\,\\ &\hat{\texttt{b5}}\texttt{*Queens[A]}\,+\, \hat{\texttt{b6}}\texttt{*Staten Island[A]}\\ \widehat{\texttt{sale_price[C]}} \;=\quad& \hat{\texttt{b0}} \,+\, \hat{\texttt{b1}}\texttt{*gross_square_feet[C]} \,+\, \hat{\texttt{b2}}\texttt{*age[C]}\,+\,\\ &\hat{\texttt{b3}}\texttt{*Bronx[C]}\,+\, \hat{\texttt{b4}}\texttt{*Brooklyn[C]} \,+\,\\ &\hat{\texttt{b5}}\texttt{*Queens[C]}\,+\, \hat{\texttt{b6}}\texttt{*Staten Island[C]} \end{align} \]
\[ \begin{align}\Leftrightarrow\qquad&\widehat{\texttt{sale_price[A]}} \,-\, \widehat{\texttt{sale_price[C]}}\qquad \\ \;=\quad &\hat{\texttt{b3}}\texttt{*}\texttt{Bronx[A]} \\ \;=\quad &\hat{\texttt{b3}}\qquad\qquad\qquad\qquad\quad\;\;\;\,\end{align} \]
Coefficient Plots in Python
- A coefficient plot shows beta estimates and their confidence intervals.
import matplotlib.pyplot as plt
import pandas as pd
# Create a Pandas DataFrame from model3's summary information
terms = assembler3.getInputCols()
coefs = model3.coefficients.toArray()[:len(terms)]
stdErrs = model3.summary.coefficientStandardErrors[:len(terms)]
df_summary = pd.DataFrame({
"term": terms,
"estimate": coefs,
"std_error": stdErrs
})
# Plot using the DataFrame columns
plt.errorbar(df_summary["term"], df_summary["estimate"],
yerr = 1.96 * df_summary["std_error"], fmt='o', capsize=5)
plt.xlabel("Terms")
plt.ylabel("Coefficient Estimate")
plt.title("Coefficient Estimates (Model 2)")
plt.axhline(0, color="red", linestyle="--") # Add horizontal line at 0
plt.xticks(rotation=45)
plt.show()Linear Regression with Log Transformation
Linear Regression with Log Transformation
The model equation with log-transformed \(\texttt{sale.price[i]}\) is \[\begin{align} \log(\texttt{sale.price[i]}) \;=\;\, &\texttt{b0} \,+\,\\ &\texttt{b1*gross.square.feet[i]} \,+\,\texttt{b2*age[i]}\,+\,\\ &\texttt{b3*Bronx[i]} \,+\,\texttt{b4*Brooklyn[i]} \,+\,\\&\texttt{b5*Queens[i]} \,+\,\texttt{b6*Staten Island[i]}\,+\,\\ &\texttt{e[i]}. \end{align}\]
- Note that the reference level for
borough_nameisManhattan.
Beta Estimates for Log-transformed Variables
1. gross_square_feet
- Let’s re-consider the two properties \(\texttt{A}\) and \(\texttt{B}\).
- \(\texttt{gross.square.feet[A]} = 2001\) and \(\texttt{gross.square.feet[B]} = 2000\).
- Both are in the same borough.
- Both properties’ ages are the same.
Beta Estimates for Log-transformed Variables
1. gross_square_feet
- If we apply the rule above for \(\widehat{\texttt{sale.price}}\texttt{[A]}\) and \(\widehat{\texttt{sale.price}}\texttt{[B]}\),
\[\begin{align}&\log(\widehat{\texttt{sale.price}}\texttt{[A]}) - \log(\widehat{\texttt{sale.price}}\texttt{[B]}) \\ \,=\, &\hat{\texttt{b1}}\,*\,(\texttt{gross.square.feet[A]} \,-\, \texttt{gross.square.feet[B]})\\ \,=\, &\hat{\texttt{b1}}\end{align}\]
So we can have the following: \[ \begin{align} &\Leftrightarrow\qquad\frac{\widehat{\texttt{sale.price[A]}}}{ \widehat{\texttt{sale.price[B]}}} \;=\; \texttt{exp(}\hat{\texttt{b1}}\texttt{)}\\ \quad&\Leftrightarrow\qquad\widehat{\texttt{sale.price[A]}} \;=\; \widehat{\texttt{sale.price[B]}} * \texttt{exp(}\hat{\texttt{b1}}\texttt{)} \end{align} \]
Beta Estimates for Log-transformed Variables
1. gross_square_feet
- Suppose \(\texttt{exp(}\hat{\texttt{b2}}\texttt{)} = 1.000431\).
- Then \(\widehat{\texttt{sale.price[A]}}\) is \(1.000431\times\widehat{\texttt{sale.price[B]}}\).
- All else being equal, an increase in \(\texttt{gross.square.feet}\) by one unit is associated with an increase in \(\texttt{sale.price}\) by 0.0431%.
Beta Estimates for Log-transformed Variables
2. borough_nameBronx
- Let’s re-consider the two properties \(\texttt{A}\) and \(\texttt{C}\).
Ais inBronx, andCis inManhattan.- Both
AandC’sageare the same. - Both
AandC’sgross.square.feetare the same.
Beta Estimates for Log-transformed Variables
2. borough_nameBronx
- If we apply the
log()-exp()rules for \(\widehat{\texttt{sale.price}}\texttt{[A]}\) and \(\widehat{\texttt{sale.price}}\texttt{[C]}\),
\[\begin{align}&\log(\widehat{\texttt{sale.price}}\texttt{[A]}) - \log(\widehat{\texttt{sale.price}}\texttt{[C]}) \\ \,=\, &\hat{\texttt{b3}}\,*\,(\texttt{borough_Bronx[A]} \,-\, \texttt{borough_Bronx[C]})\,=\, \hat{\texttt{b3}}\end{align}\]
So we can have the following: \[ \begin{align}&\Leftrightarrow\qquad\frac{\widehat{\texttt{sale.price[A]}}}{ \widehat{\texttt{sale.price[C]}}} \;=\; \texttt{exp(}\hat{\texttt{b3}}\texttt{)}\\ \quad&\Leftrightarrow\qquad\,\widehat{\texttt{sale.price[A]}} \;=\; \widehat{\texttt{sale.price[C]}} * \texttt{exp(}\hat{\texttt{b3}}\texttt{)} \end{align} \]
Beta Estimates for Log-transformed Variables
2. borough_nameBronx
- Suppose \(\texttt{exp(}\hat{\texttt{b3}}\texttt{)} = 0.2831691\).
- Then \(\widehat{\texttt{sale.price[A]}}\) is \(0.2831691\times\widehat{\texttt{sale.price[B]}}\).
- All else being equal, an increase in \(\texttt{borough_Bronx}\) by one unit is associated with a decrease in \(\texttt{sale.price}\) by (1 - 0.2831691) = 71.78%.
- All else being equal, being in
Bronxrelative to being inManhattanis associated with a decrease in \(\texttt{sale.price}\) by 71.78%.
Log-transformed Variables in PySpark
- We use the
log()function to add a log-transformed variable:
from pyspark.sql.functions import log
sale_df = sale_df.withColumn("log_sale_price",
log( sale_df['sale_price'] ) ) - To use a exponential function, we can use an
exp()function fromnumpy:
Linear Regression with Interaction Terms
Linear Regression with Interaction Terms
Motivation
Does the relationship between
sale.priceandgross.square.feetvary byborough_name?- How can linear regression address the question above?
Linear Regression with Interaction Terms
Model
- The linear regression with an interaction between predictors \(X_{1}\) and \(X_{2}\) are:
\[Y_{\texttt{i}} \,=\, b_{0} \,+\, b_{1}\,X_{1,\texttt{i}} \,+\, b_{2}\,X_{2,\texttt{i}} \,+\, b_{3}\,X_{1,\texttt{i}}\times \color{Red}{X_{2,\texttt{i}}} \,+\, e_{\texttt{i}},\]
- where
- \(i\;\): \(\;\;i\)-th observation in the training DataFrame, \(i = 1, 2, 3, \cdots\).
- \(Y_{i}\,\): \(\;i\)-th observation of outcome \(Y\).
- \(X_{p, i}\,\): \(i\)-th observation of the \(p\)-th predictor \(X_{p}\).
- \(e_{i}\;\): \(\;i\)-th observation of statistical error.
Linear Regression with Interaction Terms
Model
- The linear regression with an interaction between predictors \(X_{1}\) and \(X_{2}\) are:
\[Y_{\texttt{i}} \,=\, b_{0} \,+\, b_{1}\,X_{1,\texttt{i}} \,+\, b_{2}\,X_{2,\texttt{i}} \,+\, b_{3}\,X_{1,\texttt{i}}\times \color{Red}{X_{2,\texttt{i}}} \,+\, e_{\texttt{i}}\;.\]
Interaction
- The relationship between \(X_{1}\) and \(Y\) varies by values of \(b_{3}\, X_{2}\):
\[\frac{\Delta Y}{\Delta X_{1}} \,=\, b_{1} + b_{3}\, X_{2}\].
Example
- \(X_{2}\) is often a dummy variable. If \(b_{3} \neq 0\) and \(X_{2, \texttt{i}} = 1\),
\[\frac{\Delta Y}{\Delta X_{1}} \,=\, b_{1} + b_{3}\].
Linear Regression with Interaction Terms
Interaction with a Dummy Variable
- The linear regression with an interaction between predictors \(X_{1}\) and \(X_{2}\in\{\,0, 1\,\}\) are:
\[Y_{\texttt{i}} \,=\, b_{0} \,+\, b_{1}\,X_{1,\texttt{i}} \,+\, b_{2}\,X_{2,\texttt{i}} \,+\, b_{3}\,X_{1,\texttt{i}}\times \color{Red}{X_{2,\texttt{i}}} \,+\, e_{\texttt{i}},\] where \(X_{\,2, \texttt{i}}\) is either 0 or 1.
For \(\texttt{i}\) such that \(X_{\,2, \texttt{i}} = 0\), the model is \[Y_{\texttt{i}} \,=\, b_{0} \,+\, b_{1}\,X_{1,\texttt{i}} \,+\, e_{\texttt{i}}\qquad\qquad\qquad\qquad\qquad\quad\;\;\]
For \(\texttt{i}\) such that \(X_{\,2, \texttt{i}} = 1\), the model is
\[Y_{\texttt{i}} \,=\, (\,b_{0} \,+\, b_{2}\,) \,+\, (\,b_{1}\,+\, b_{3}\,)\,X_{1,\texttt{i}} \,+\, e_{\texttt{i}}\qquad\qquad\]
Linear Regression with Interaction Terms
Motivation
- Is
sale.pricerelated withgross.square.feet?
\[ \begin{align} \texttt{sale_price[i]} \;=\;\, &\texttt{b0} \,+\,\\ &\texttt{b1*Bronx[i]} \,+\,\texttt{b2*Brooklyn[i]} \,+\,\\&\texttt{b3*Queens[i]} \,+\,\texttt{b4*Staten Island[i]}\,+\,\\ &\texttt{b5*age[i]}\,+\,\\ &\texttt{b6*gross_square_feet[i]} \,+\,\texttt{e[i]} \end{align} \]
Linear Regression with Interaction Terms
Motivation
- Does the relationship between
sale.priceandgross.square.feetvary byborough_name?
\[ \begin{align} \texttt{sale_price[i]} \;=\;\, &\texttt{b0} \,+\,\\ &\texttt{b1*Bronx[i]} \,+\,\texttt{b2*Brooklyn[i]} \,+\,\\ &\texttt{b3*Queens[i]} \,+\,\texttt{b4*Staten Island[i]}\,+\, \\ &\texttt{b5*age[i]}\,+\,\\ &\texttt{b6*gross_square_feet[i]} \,+\,\\ &\texttt{b7*gross_square_feet[i]*Bronx[i]} \,+\, \\ &\texttt{b8*gross_square_feet[i]*Brooklyn[i]} \,+\, \\ &\texttt{b9*gross_square_feet[i]*Queens[i]} \,+\, \\ &\texttt{b10*gross_square_feet[i]*Staten Island[i]} \,+\, \texttt{e[i]} \\ \end{align} \]
Log-Log Linear Regression
Log-Log Linear Regression
- To estimate the price elasticity of orange juice (OJ), we will use sales data for OJ from Dominick’s grocery stores in the 1990s.
- Weekly
priceandsales(in number of cartons “sold”) for three OJ brandsTropicana, Minute Maid, Dominick’s - A dummy,
ad, showing whether eachbrandwas advertised (in store or flyer) that week.
- Weekly
| Variable | Description |
|---|---|
sales |
Quantity of OJ cartons sold |
price |
Price of OJ |
brand |
Brand of OJ |
ad |
Advertisement status |
Log-Log Linear Regression
Estimating Price Elasticity
- Let’s prepare the OJ data:
Log-Log Linear Regression
Estimating Price Elasticity
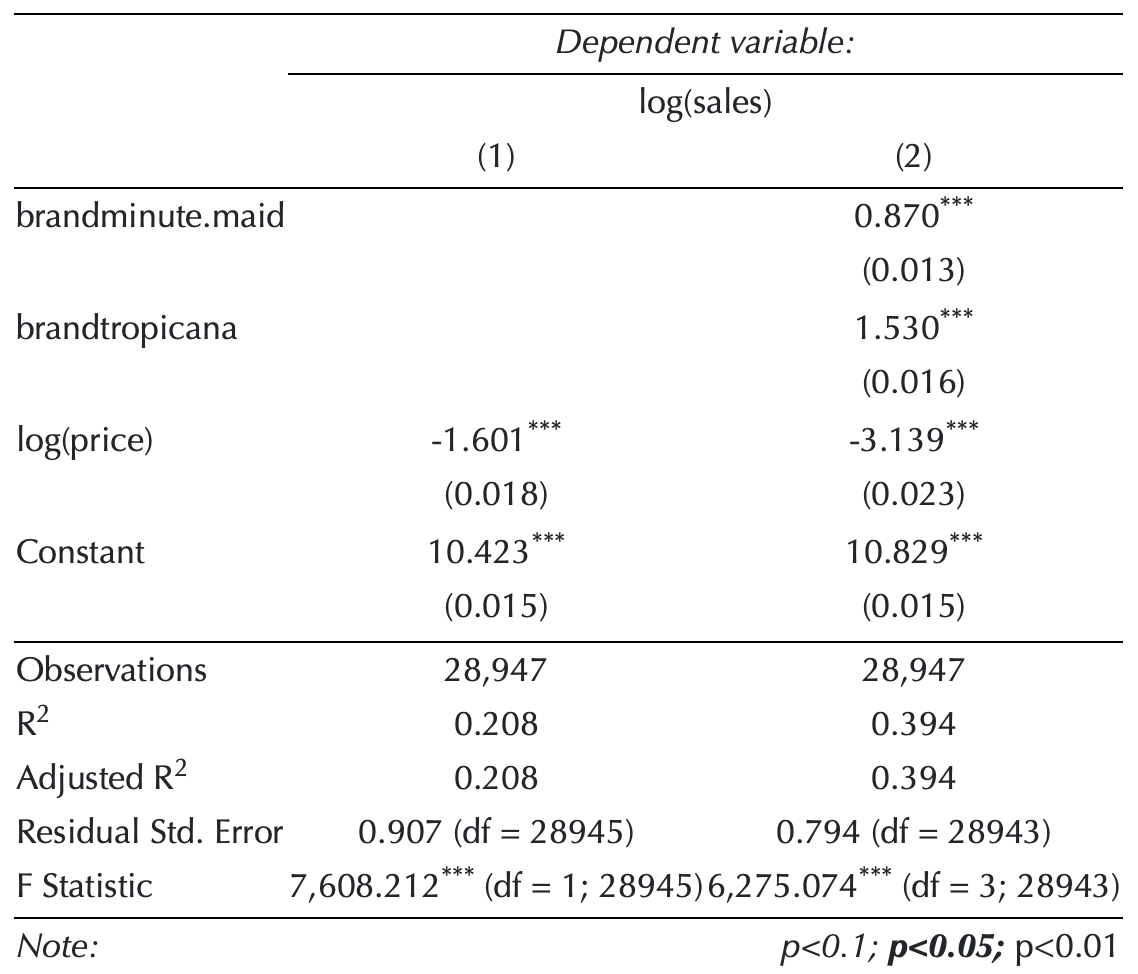
- The following model estimates the price elasticity of demand for a carton of OJ:
\[\begin{align} \log(\texttt{sales}_{\texttt{i}}) &\,=\, \;\; b_{\texttt{intercept}} \,+\, b_{\,\texttt{mm}}\,\texttt{brand}_{\,\texttt{mm}, \texttt{i}} \,+\, b_{\,\texttt{tr}}\,\texttt{brand}_{\,\texttt{tr}, \texttt{i}}\\ &\quad\,+\, b_{\texttt{price}}\,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}},\\ \text{where}\qquad\qquad&\\ \texttt{brand}_{\,\texttt{tr}, \texttt{i}} &\,=\, \begin{cases} \texttt{1} & \text{ if an orange juice } \texttt{i} \text{ is } \texttt{Tropicana};\\\\ \texttt{0} & \text{otherwise}.\qquad\qquad\quad\, \end{cases}\\ \texttt{brand}_{\,\texttt{mm}, \texttt{i}} &\,=\, \begin{cases} \texttt{1} & \text{ if an orange juice } \texttt{i} \text{ is } \texttt{Minute Maid};\\\\ \texttt{0} & \text{otherwise}.\qquad\qquad\quad\, \end{cases} \end{align}\]
Log-Log Linear Regression
Estimating Price Elasticity
- The following model estimates the price elasticity of demand for a carton of OJ:
\[\log(\texttt{sales}_{\texttt{i}}) \,=\, \quad\;\; b_{\texttt{intercept}} \,+\, b_{\,\texttt{mm}}\,\texttt{brand}_{\,\texttt{mm}, \texttt{i}} \,+\, b_{\,\texttt{tr}}\,\texttt{brand}_{\,\texttt{tr}, \texttt{i}}\\ \,+\, b_{\texttt{price}}\,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}}\]
When \(\texttt{brand}_{\,\texttt{tr}, \texttt{i}}\,=\,0\) and \(\texttt{brand}_{\,\texttt{mm}, \texttt{i}}\,=\,0\), the beta coefficient for the intercept \(b_{\texttt{intercept}}\) gives the value of Dominick’s log sales at \(\log(\,\texttt{price[i]}\,) = 0\).
The beta coefficient \(b_{\texttt{price}}\) is the price elasticity of demand.
- It measures how sensitive the quantity demanded is to its price.
Log-Log Linear Regression
Estimating Price Elasticity
- For small changes in variable \(x\) from \(x_{0}\) to \(x_{1}\), the following equation holds:
\[\Delta \log(x) \,= \, \log(x_{1}) \,-\, \log(x_{0}) \approx\, \frac{x_{1} \,-\, x_{0}}{x_{0}} \,=\, \frac{\Delta\, x}{x_{0}}.\]
- The coefficient on \(\log(\texttt{price}_{\texttt{i}})\), \(b_{\texttt{price}}\), is therefore
\[b_{\texttt{price}} \,=\, \frac{\Delta \log(\texttt{sales}_{\texttt{i}})}{\Delta \log(\texttt{price}_{\texttt{i}})}\,=\, \frac{\frac{\Delta \texttt{sales}_{\texttt{i}}}{\texttt{sales}_{\texttt{i}}}}{\frac{\Delta \texttt{price}_{\texttt{i}}}{\texttt{price}_{\texttt{i}}}}.\]
- All else being equal, an increase in \(\texttt{price}\) by 1% is associated with a decrease in \(\texttt{sales}\) by \(b_{\texttt{price}}\)%.
Log-Log Linear Regression
EDA
- Describe the relationship between
log(price)andlog(sales)bybrand.
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
oj['log_price'] = oj['price'].apply(lambda x: np.log(x))
oj['log_sales'] = oj['sales'].apply(lambda x: np.log(x))
sns.lmplot(
x='log_price',
y='log_sales',
hue='brand', # color by brand
data=oj,
aspect=1.2,
height=5,
markers=["o", "s", "D"], # optional: different markers per brand
scatter_kws={'alpha': 0.025}, # Pass alpha to the scatter plot
legend=True
)Log-Log Linear Regression
EDA
- Describe the relationship between
log(price)andlog(sales)bybrandandad.
sns.lmplot(
x='log_price',
y='log_sales',
hue='brand', # color by brand
col='ad', # facet by feat
data=oj,
aspect=1.2,
height=5,
markers=["o", "s", "D"], # optional: different markers per brand
# Pass alpha to the scatter plot
scatter_kws={'alpha': 0.025},
col_wrap=2, # how many facets per row
legend=True
)Log-Log Linear Regression
Estimating Price Elasticity
- Let’s train the first model,
model_1:
\[ \begin{align} \log(\texttt{sales}_{\texttt{i}}) \,=\, &\quad\;\; b_{\texttt{intercept}} \\ &\,+\,b_{\,\texttt{mm}}\,\texttt{brand}_{\,\texttt{mm}, \texttt{i}} \,+\, b_{\,\texttt{tr}}\,\texttt{brand}_{\,\texttt{tr}, \texttt{i}}\\ &\,+\, b_{\texttt{price}}\,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}} \end{align} \]
Log-Log Linear Regression
Estimating Price Elasticity
How does the relationship between
log(sales)andlog(price)vary bybrand?- Let’s train the second model,
model_2, that addresses the above question:
- Let’s train the second model,
\[ \begin{align} \log(\texttt{sales}_{\texttt{i}}) \,=\,&\;\; \quad b_{\texttt{intercept}} \,+\, \color{Green}{b_{\,\texttt{mm}}\,\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} \,+\, \color{Blue}{b_{\,\texttt{tr}}\,\texttt{brand}_{\,\texttt{tr}, \texttt{i}}}\\ &\,+\, b_{\texttt{price}}\,\log(\texttt{price}_{\texttt{i}}) \\ &\, +\, b_{\texttt{price*mm}}\,\log(\texttt{price}_{\texttt{i}})\,\times\,\color{Green} {\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} \\ &\,+\, b_{\texttt{price*tr}}\,\log(\texttt{price}_{\texttt{i}})\,\times\,\color{Blue} {\texttt{brand}_{\,\texttt{tr}, \texttt{i}}} \,+\, e_{\texttt{i}} \end{align} \]
Log-Log Linear Regression
For \(\texttt{i}\) such that \(\color{Green}{\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} = 0\) and \(\color{Blue}{\texttt{brand}_{\,\texttt{tr}, \texttt{i}}} = 0\), the model equation is: \[\log(\texttt{sales}_{\texttt{i}}) \,=\, \; \,b_{\texttt{intercept}}\qquad\qquad\qquad\qquad\qquad\qquad\qquad\qquad\\ \qquad\,+\, b_{\texttt{price}} \,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}}\,.\qquad\qquad\;\]
For \(\texttt{i}\) such that \(\color{Green}{\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} = 1\) and \(\color{Blue}{\texttt{brand}_{\,\texttt{tr}, \texttt{i}}} = 0\), the model equation is: \[\log(\texttt{sales}_{\texttt{i}}) \,=\, \; (\,b_{\texttt{intercept}} \,+\, b_{\,\texttt{mm}}\,)\qquad\qquad\qquad\qquad\qquad\qquad\qquad\\ \qquad\!\,+\,(\, b_{\texttt{price}} \,+\, \color{Green}{b_{\texttt{price*mm}}}\,)\,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}}\,.\]
For \(\texttt{i}\) such that \(\color{Green}{\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} = 0\) and \(\color{Blue}{\texttt{brand}_{\,\texttt{tr}, \texttt{i}}} = 1\), the model equation is: \[\log(\texttt{sales}_{\texttt{i}}) \,=\, \; (\,b_{\texttt{intercept}} \,+\, b_{\,\texttt{tr}}\,)\qquad\qquad\qquad\qquad\qquad\qquad\qquad\\ \qquad\!\,+\,(\, b_{\texttt{price}} \,+\, \color{Blue}{b_{\texttt{price*tr}}}\,)\,\log(\texttt{price}_{\texttt{i}}) \,+\, e_{\texttt{i}}\,.\]
Adding Interaction Terms in PySpark
The UDF
add_interaction_termsrequires at least two lists of variable names.var_list3is optional.- If
var_list3is provided, theadd_interaction_termsfunction adds all possible two-way interactions and three-way interaction to thedtrainanddtestDataFrames.
- If
def add_interaction_terms(var_list1, var_list2, var_list3=None):
"""
Creates interaction term columns in the global DataFrames dtrain and dtest.
For two sets of variable names (which may represent categorical (dummy) or continuous variables),
this function creates two-way interactions by multiplying each variable in var_list1 with each
variable in var_list2.
Optionally, if a third list of variable names (var_list3) is provided, the function also creates
three-way interactions among each variable in var_list1, each variable in var_list2, and each variable
in var_list3.
Parameters:
var_list1 (list): List of column names for the first set of variables.
var_list2 (list): List of column names for the second set of variables.
var_list3 (list, optional): List of column names for the third set of variables for three-way interactions.
Returns:
A flat list of new interaction column names.
"""
global dtrain, dtest
interaction_cols = []
# Create two-way interactions between var_list1 and var_list2.
for var1 in var_list1:
for var2 in var_list2:
col_name = f"{var1}_*_{var2}"
dtrain = dtrain.withColumn(col_name, col(var1).cast("double") * col(var2).cast("double"))
dtest = dtest.withColumn(col_name, col(var1).cast("double") * col(var2).cast("double"))
interaction_cols.append(col_name)
# Create two-way interactions between var_list1 and var_list3.
if var_list3 is not None:
for var1 in var_list1:
for var3 in var_list3:
col_name = f"{var1}_*_{var3}"
dtrain = dtrain.withColumn(col_name, col(var1).cast("double") * col(var3).cast("double"))
dtest = dtest.withColumn(col_name, col(var1).cast("double") * col(var3).cast("double"))
interaction_cols.append(col_name)
# Create two-way interactions between var_list2 and var_list3.
if var_list3 is not None:
for var2 in var_list2:
for var3 in var_list3:
col_name = f"{var2}_*_{var3}"
dtrain = dtrain.withColumn(col_name, col(var2).cast("double") * col(var3).cast("double"))
dtest = dtest.withColumn(col_name, col(var2).cast("double") * col(var3).cast("double"))
interaction_cols.append(col_name)
# If a third list is provided, create three-way interactions.
if var_list3 is not None:
for var1 in var_list1:
for var2 in var_list2:
for var3 in var_list3:
col_name = f"{var1}_*_{var2}_*_{var3}"
dtrain = dtrain.withColumn(col_name, col(var1).cast("double") * col(var2).cast("double") * col(var3).cast("double"))
dtest = dtest.withColumn(col_name, col(var1).cast("double") * col(var2).cast("double") * col(var3).cast("double"))
interaction_cols.append(col_name)
return interaction_cols
# Example
# interaction_cols_brand_price = add_interaction_terms(dummy_cols_brand, ['log_price'])
# interaction_cols_brand_ad_price = add_interaction_terms(dummy_cols_brand, dummy_cols_ad, ['log_price'])Log-Log Linear Regression
- How does advertisement play a role in the relationship between sales and prices in the OJ market?
- The ads can increase sales at all prices.
- They can change price sensitivity.
- They can do both of these things in a brand-specific manner.
- What would be the formula that address the question above?
Log-Log Linear Regression
- Let’s train the third model,
model_3, that addresses the question in the previous page:
\[ \begin{align} \log(\texttt{sales}_{\texttt{i}}) \,=\,\quad\;\;& b_{\texttt{intercept}} \,+\, \color{Green}{b_{\,\texttt{mm}}\,\texttt{brand}_{\,\texttt{mm}, \texttt{i}}} \,+\, \color{Blue}{b_{\,\texttt{tr}}\,\texttt{brand}_{\,\texttt{tr}, \texttt{i}}} \\ &\,+\; b_{\,\texttt{ad}}\,\color{Orange}{\texttt{ad}_{\,\texttt{i}}} \qquad\qquad\qquad\qquad\quad \\ &\,+\, b_{\texttt{mm*ad}}\,\color{Green} {\texttt{brand}_{\,\texttt{mm}, \texttt{i}}}\,\times\, \color{Orange}{\texttt{ad}_{\,\texttt{i}}}\,+\, b_{\texttt{tr*ad}}\,\color{Blue} {\texttt{brand}_{\,\texttt{tr}, \texttt{i}}}\,\times\, \color{Orange}{\texttt{ad}_{\,\texttt{i}}} \\ &\,+\; b_{\texttt{price}}\,\log(\texttt{price}_{\texttt{i}}) \qquad\qquad\qquad\;\;\;\;\, \\ &\,+\, b_{\texttt{price*mm}}\,\log(\texttt{price}_{\texttt{i}})\,\times\,\color{Green} {\texttt{brand}_{\,\texttt{mm}, \texttt{i}}}\qquad\qquad\qquad\;\, \\ &\,+\, b_{\texttt{price*tr}}\,\log(\texttt{price}_{\texttt{i}})\,\times\,\color{Blue} {\texttt{brand}_{\,\texttt{tr}, \texttt{i}}}\qquad\qquad\qquad\;\, \\ & \,+\, b_{\texttt{price*ad}}\,\log(\texttt{price}_{\texttt{i}})\,\times\,\color{Orange}{\texttt{ad}_{\,\texttt{i}}}\qquad\qquad\qquad\;\;\, \\ &\,+\, b_{\texttt{price*mm*ad}}\,\log(\texttt{price}_{\texttt{i}}) \,\times\,\,\color{Green} {\texttt{brand}_{\,\texttt{mm}, \texttt{i}}}\,\times\, \color{Orange}{\texttt{ad}_{\,\texttt{i}}} \\ &\,+\, b_{\texttt{price*tr*ad}}\,\log(\texttt{price}_{\texttt{i}}) \,\times\,\,\color{Blue} {\texttt{brand}_{\,\texttt{tr}, \texttt{i}}}\,\times\, \color{Orange}{\texttt{ad}_{\,\texttt{i}}} \,+\, e_{\texttt{i}} \end{align} \]
Log-Log Linear Regression
Estimating Price Elasticity
- Describe the relationship between
adandbrandusing stacked bar charts.
sns.histplot(
data=oj,
x='ad_status', # categorical variable on the x-axis
hue='brand', # fill color by brand
multiple='fill' # 100% stacked bars
)
plt.ylabel('Proportion')- Model 3 assumes that the relationship between
priceandsalescan vary byad.
Log-Log Linear Regression
Estimating Price Elasticity
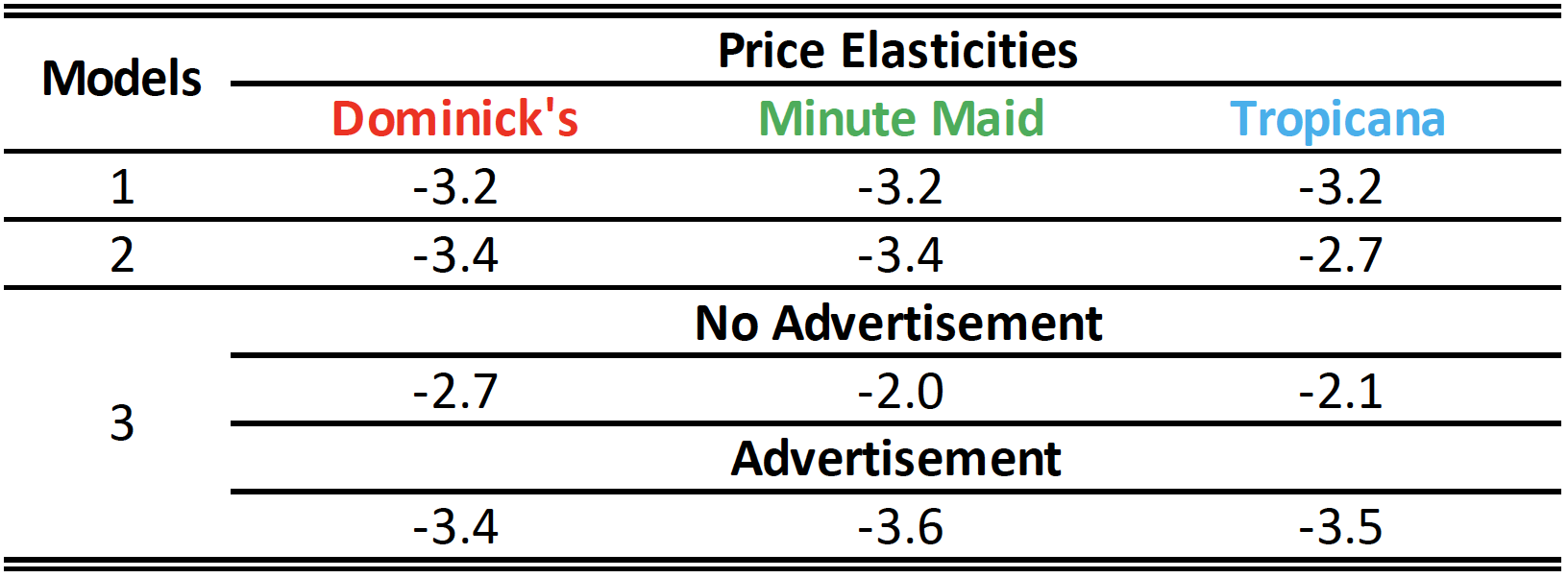
How would you explain different estimation results across different models?
Which model do you prefer? Why?